You are browsing environment: FUNGIDB
CAZyme Information: OAG24480.1
You are here: Home > Sequence: OAG24480.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG24480.1 | |||||||||||
| CAZy Family | GT15 | |||||||||||
| CAZyme Description | mixed-linked glucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.4:107 | 3.2.1.151:22 | 3.2.1.73:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH12 | 90 | 236 | 8.1e-47 | 0.9807692307692307 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396303 | Glyco_hydro_12 | 1.42e-74 | 32 | 235 | 1 | 204 | Glycosyl hydrolase family 12. |
| 235746 | PRK06215 | 1.93e-22 | 8 | 237 | 26 | 237 | hypothetical protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.76e-149 | 1 | 238 | 1 | 239 | |
| 6.52e-145 | 1 | 238 | 1 | 243 | |
| 3.09e-104 | 11 | 236 | 16 | 243 | |
| 7.55e-96 | 6 | 235 | 4 | 244 | |
| 7.55e-96 | 6 | 235 | 4 | 244 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-94 | 18 | 236 | 5 | 225 | Chain A, glycoside hydrolase family 12 beta-1,3-1,4-glucanase [Chaetomium sp.],7EEE_A Chain A, glycoside hydrolase family 12 beta-1,3-1,4-glucanase [Chaetomium sp.],7EEJ_A Chain A, glycoside hydrolase family 12 beta-1,3-1,4-glucanase [Chaetomium sp.] |
|
| 1.12e-88 | 7 | 234 | 5 | 231 | CRYSTAL STRUCTURE 4Ac Endoglucanase-like protein from Acremonium chrysogenum [Acremonium chrysogenum ATCC 11550],5M2D_B CRYSTAL STRUCTURE 4Ac Endoglucanase-like protein from Acremonium chrysogenum [Acremonium chrysogenum ATCC 11550] |
|
| 1.02e-85 | 22 | 236 | 3 | 217 | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 [Aspergillus aculeatus],5GM3_B Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 [Aspergillus aculeatus] |
|
| 4.26e-85 | 19 | 236 | 1 | 218 | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose [Aspergillus aculeatus],5GM5_B Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose [Aspergillus aculeatus],5GM5_C Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose [Aspergillus aculeatus],5GM5_D Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose [Aspergillus aculeatus],5GM5_E Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose [Aspergillus aculeatus],5GM5_F Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose [Aspergillus aculeatus],5GM5_G Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose [Aspergillus aculeatus] |
|
| 8.29e-85 | 22 | 236 | 3 | 217 | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose [Aspergillus aculeatus],5GM4_B Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose [Aspergillus aculeatus],5GM4_C Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose [Aspergillus aculeatus],5GM4_D Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose [Aspergillus aculeatus],5GM4_E Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose [Aspergillus aculeatus],5GM4_F Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose [Aspergillus aculeatus],5GM4_G Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose [Aspergillus aculeatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.32e-89 | 7 | 236 | 6 | 235 | Endoglucanase-1 OS=Aspergillus aculeatus OX=5053 PE=1 SV=1 |
|
| 4.74e-83 | 4 | 236 | 3 | 237 | Endoglucanase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=cekA PE=2 SV=2 |
|
| 2.12e-78 | 18 | 236 | 35 | 255 | Endoglucanase cel12A OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel12A PE=1 SV=1 |
|
| 3.64e-68 | 21 | 235 | 37 | 260 | Endoglucanase cel12B OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel12B PE=1 SV=1 |
|
| 1.78e-57 | 1 | 236 | 1 | 237 | Xyloglucan-specific endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgeA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
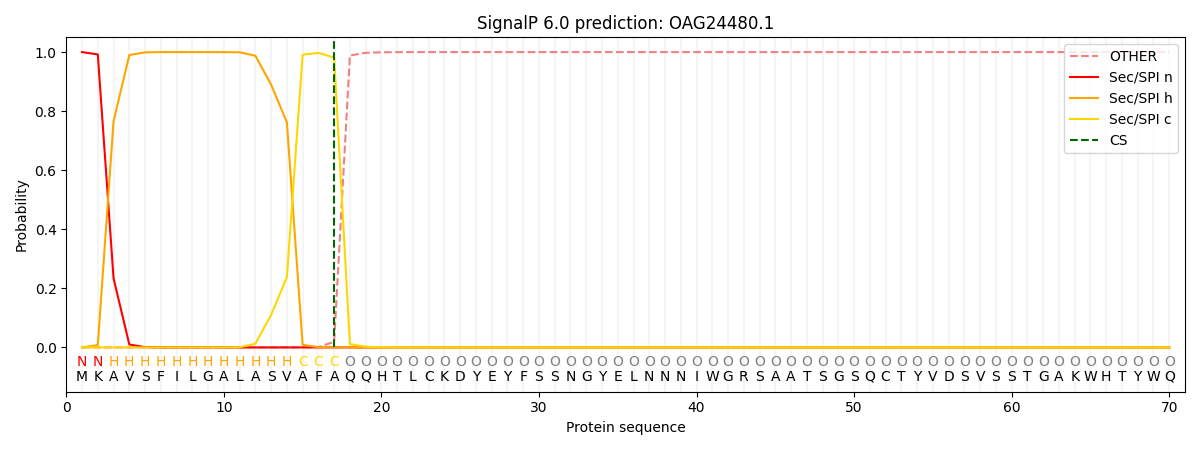
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000228 | 0.999768 | CS pos: 17-18. Pr: 0.9812 |
