You are browsing environment: FUNGIDB
CAZyme Information: OAG24208.1
You are here: Home > Sequence: OAG24208.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG24208.1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | 1,3-beta-glucan synthase component GLS2 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 75344; End:81297 Strand: + | |||||||||||
Full Sequence Download help
| MSGHPQGGHY DDGYGQTQGH AQGQQGQDAY YHDDQYSQYH DDAHGAQYQQ QGQHGDAYYD | 60 |
| EQGYYDGQQQ GGHYQQNGYY DDRGQQGYQD EYYNDQYYDQ GGAQDGYAAN GQQPRRRQHD | 120 |
| SEEDSETFSD FTMRSDMARA TDMDYYGRGD ERYNSYAEGN RGYRPPSSQV SYGGNRSSGA | 180 |
| STPIYGMDYS NALPAGQRSR EPYPAWTAEA QIPCTKEEIE DIFLDLTAKF GFQRDSMRNM | 240 |
| YDHFMTLLDS RASRMSPNQA LLSLHADYIG GENANYRRWY FAAHLDLDDA VGFANMNLGK | 300 |
| ASRRTRKARK AAKKKASENP GNEQETLDAY EGDNSLEAAE YRWKTRMNRM SQHDRVRQVA | 360 |
| LYLLCWGEAN QVRFMPELMC FIFKCADDWL NSPAGQAQTE PIEEFTYLNQ VITPLYQYCR | 420 |
| DQGYEIQDGK YVRRERDHAA IIGYDDMNQL FWYPEGLERI VFEDKSRLVD IPPAERYLKL | 480 |
| KDVLWKKVFF KTYYERRSWF HMVINFNRIW VIHLTSFWFY TAYNSQPLYT KNYQQQLDQK | 540 |
| PEKAATLSAV ALGGTIAAFI QIFATICEWC YVPRRWAGAQ HLTKRLLFLI AVFVVNIAPS | 600 |
| VYIFGLDKRT GTIANVLGGV QFAIALVTFI FFSLMPIGGL FGSYLTRNSR KYVASQTFTA | 660 |
| SYPRLNGNDK WMSYGLWVLV FAAKLAESYF FLTLSIKDPI RILSHMKKPD CLGDAIIGNI | 720 |
| LCKYQPRILL GLMYFMDLIL FFLDSYLWYI IANMLFSVSR SFYLGVSIWT PWRNIFSRLP | 780 |
| KRIYSKVLAT TDMEIKYKPK VLISQIWNAV VISMYREHLL AIDHVQKLLY HQVPSEQEGK | 840 |
| RTLRAPTFFV SQEDHSFKTE FFPAQSEAER RISFFAQSLS TPIPEPLPVD NMPTFTVLIP | 900 |
| HYGEKILLSL REIIREDEPY SRVTLLEYLK QLHPHEWDCF VKDTKILADE TSQFNGDDEK | 960 |
| GEKDTAKSKI DDLPFYCIGF KSAAPEYTLR TRIWASLRSQ TLYRTISGFM NYSRAIKLLY | 1020 |
| RVENPEVVQM FGGNSDKLER ELERMARRKY KICVSMQRYA KFNTEERENT EFLLRAYPDL | 1080 |
| QIAYLDEEPP ENEGEEPRIY SALIDGHSEI MDNGMRRPKF RIQLSGNPIL GDGKSDNQNH | 1140 |
| SIIFYRGEYI QLIDANQDNY LEECLKIRSV LAEFEEMTTD NVSPYTPGLP STKFNPVAIL | 1200 |
| GAREYIFSEN IGILGDIAAG KEQTFGTMFA RTLAQIGGKL HYGHPDFLNG IFMTTRGGVS | 1260 |
| KAQKGLHLNE DIYAGMNALL RGGRIKHCEY YQCGKGRDLG FGSVLNFTTK IGTGMGEQML | 1320 |
| SREYYYMGTQ LPLDRFLSFY YAHPGFHINN MFIMLSVQCF MFVLLNLGAL NHETILCQFN | 1380 |
| KDIPITDPQW PNGCANLVPV FDWVSRCIVS IFIVFFISFV PLVVQELTER GFWRAATRLA | 1440 |
| KHFSSGSPFF EVFVTQIYSN ALHTNLSYGG ARYIGTGRGF ATARIPFGIL FSRFAGPSIY | 1500 |
| IGARSLMMLL FATITVWGPW LIYFWASLMS LCLAPFLFNP HQFSWDDFFI DYREYLRWLS | 1560 |
| RGNTRSHSAS WIGYCRLSRT RITGFKRKIL GDPSAKLSGD VPRAHFTNIF MSEILGPLVL | 1620 |
| VAITLIPFLF INAQTGVNDN RHKDDDTATG DPKKSGALVR IAIIAFGPVA INAGVLGGMF | 1680 |
| ALACCAGPLL SMCCKKFGAV LAAIAHAIAV LVLLIFFVVL MFLEGFSFPR ALSGMIAVVA | 1740 |
| IQRFVFKLII TLALTREFKA DTANIAWWTG KWYTMGWHTI SQPGREFLCK ITELGFFAAD | 1800 |
| FILGHVLLFF MLPILLIPYA DKGHSMMLFW LRPSRQIRPP IYSLKQTKLR KRRVVRYAIL | 1860 |
| YFVLLVVFLA LIVGPIVAGK YIKLSVTLPM EILQPTGFSN NDTTNTPTGK CVDERPCPVW | 1920 |
| NPAVDATGGG GAAASTDDAA RRFRRYMAY | 1949 |
Enzyme Prediction help
| EC | 2.4.1.34:56 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 861 | 1640 | 0 | 0.9905277401894452 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 861 | 1690 | 1 | 819 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 5.45e-47 | 354 | 463 | 1 | 111 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 367362 | Glutenin_hmw | 0.002 | 47 | 205 | 271 | 417 | High molecular weight glutenin subunit. Members of this family include high molecular weight subunits of glutenin. This group of gluten proteins is thought to be largely responsible for the elastic properties of gluten, and hence, doughs. Indeed, glutenin high molecular weight subunits are classified as elastomeric proteins, because the glutenin network can withstand significant deformations without breaking, and return to the original conformation when the stress is removed. Elastomeric proteins differ considerably in amino acid sequence, but they are all polymers whose subunits consist of elastomeric domains, composed of repeated motifs, and non-elastic domains that mediate cross-linking between the subunits. The elastomeric domain motifs are all rich in glycine residues in addition to other hydrophobic residues. High molecular weight glutenin subunits have an extensive central elastomeric domain, flanked by two terminal non-elastic domains that form disulphide cross-links. The central elastomeric domain is characterized by the following three repeated motifs: PGQGQQ, GYYPTS[P/L]QQ, GQQ. It possesses overlapping beta-turns within and between the repeated motifs, and assumes a regular helical secondary structure with a diameter of approx. 1.9 nm and a pitch of approx. 1.5 nm. |
| 367362 | Glutenin_hmw | 0.004 | 2 | 202 | 290 | 469 | High molecular weight glutenin subunit. Members of this family include high molecular weight subunits of glutenin. This group of gluten proteins is thought to be largely responsible for the elastic properties of gluten, and hence, doughs. Indeed, glutenin high molecular weight subunits are classified as elastomeric proteins, because the glutenin network can withstand significant deformations without breaking, and return to the original conformation when the stress is removed. Elastomeric proteins differ considerably in amino acid sequence, but they are all polymers whose subunits consist of elastomeric domains, composed of repeated motifs, and non-elastic domains that mediate cross-linking between the subunits. The elastomeric domain motifs are all rich in glycine residues in addition to other hydrophobic residues. High molecular weight glutenin subunits have an extensive central elastomeric domain, flanked by two terminal non-elastic domains that form disulphide cross-links. The central elastomeric domain is characterized by the following three repeated motifs: PGQGQQ, GYYPTS[P/L]QQ, GQQ. It possesses overlapping beta-turns within and between the repeated motifs, and assumes a regular helical secondary structure with a diameter of approx. 1.9 nm and a pitch of approx. 1.5 nm. |
| 367362 | Glutenin_hmw | 0.007 | 35 | 205 | 178 | 342 | High molecular weight glutenin subunit. Members of this family include high molecular weight subunits of glutenin. This group of gluten proteins is thought to be largely responsible for the elastic properties of gluten, and hence, doughs. Indeed, glutenin high molecular weight subunits are classified as elastomeric proteins, because the glutenin network can withstand significant deformations without breaking, and return to the original conformation when the stress is removed. Elastomeric proteins differ considerably in amino acid sequence, but they are all polymers whose subunits consist of elastomeric domains, composed of repeated motifs, and non-elastic domains that mediate cross-linking between the subunits. The elastomeric domain motifs are all rich in glycine residues in addition to other hydrophobic residues. High molecular weight glutenin subunits have an extensive central elastomeric domain, flanked by two terminal non-elastic domains that form disulphide cross-links. The central elastomeric domain is characterized by the following three repeated motifs: PGQGQQ, GYYPTS[P/L]QQ, GQQ. It possesses overlapping beta-turns within and between the repeated motifs, and assumes a regular helical secondary structure with a diameter of approx. 1.9 nm and a pitch of approx. 1.5 nm. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEL31281.1|GT48 | 0.0 | 1 | 1949 | 1 | 1951 |
| CAE7209855.1|GT48 | 0.0 | 1 | 1949 | 1 | 1942 |
| CBX98352.1|GT48 | 0.0 | 1 | 1949 | 1 | 1947 |
| QRD01344.1|GT48 | 0.0 | 1 | 1949 | 1 | 1950 |
| ABL63820.1|GT48 | 0.0 | 1 | 1912 | 1 | 1891 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P40989|FKS2_YEAST | 0.0 | 108 | 1903 | 83 | 1870 | 1,3-beta-glucan synthase component GSC2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GSC2 PE=1 SV=2 |
| sp|A2QLK4|FKS1_ASPNC | 0.0 | 55 | 1909 | 54 | 1878 | 1,3-beta-glucan synthase component FKS1 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=fksA PE=3 SV=1 |
| sp|Q04952|FKS3_YEAST | 0.0 | 203 | 1908 | 15 | 1777 | 1,3-beta-glucan synthase component FKS3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS3 PE=1 SV=1 |
| sp|P38631|FKS1_YEAST | 0.0 | 111 | 1903 | 58 | 1851 | 1,3-beta-glucan synthase component FKS1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS1 PE=1 SV=2 |
| sp|O93927|FKS1_CRYNH | 0.0 | 165 | 1877 | 104 | 1781 | 1,3-beta-glucan synthase component FKS1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=FKS1 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000054 | 0.000004 |
TMHMM Annotations download full data without filtering help
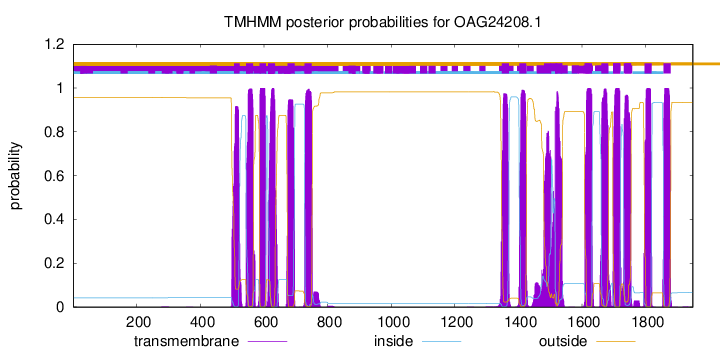
| Start | End |
|---|---|
| 506 | 523 |
| 544 | 566 |
| 586 | 606 |
| 613 | 635 |
| 674 | 696 |
| 728 | 750 |
| 1348 | 1370 |
| 1403 | 1425 |
| 1481 | 1503 |
| 1516 | 1538 |
| 1609 | 1631 |
| 1661 | 1683 |
| 1698 | 1720 |
| 1732 | 1754 |
| 1797 | 1819 |
| 1857 | 1879 |
