You are browsing environment: FUNGIDB
OAG22830.1
Basic Information
help
Species
Alternaria alternata
Lineage
Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata
CAZyme ID
OAG22830.1
CAZy Family
GH55
CAZyme Description
glycoside hydrolase
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
320
35240.06
6.2295
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_AalternataSRC1lrK2f
13577
N/A
111
13466
Gene Location
Family
Start
End
Evalue
family coverage
GH30
1
313
7.5e-96
0.7192982456140351
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
405300
Glyco_hydr_30_2
3.98e-10
1
183
132
353
O-Glycosyl hydrolase family 30.
more
396577
Glyco_hydro_59
1.53e-05
66
183
138
251
Glycosyl hydrolase family 59.
more
227807
XynC
0.002
1
313
129
426
O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis].
more
399741
Alpha-L-AF_C
0.005
198
275
63
152
Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.98e-191
1
320
161
480
1.55e-186
1
320
162
482
6.41e-164
2
320
162
481
5.54e-155
1
320
160
480
5.54e-155
1
320
160
480
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
7.68e-09
1
261
94
341
Crystal Structure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
6.65e-118
1
320
160
478
Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1
more
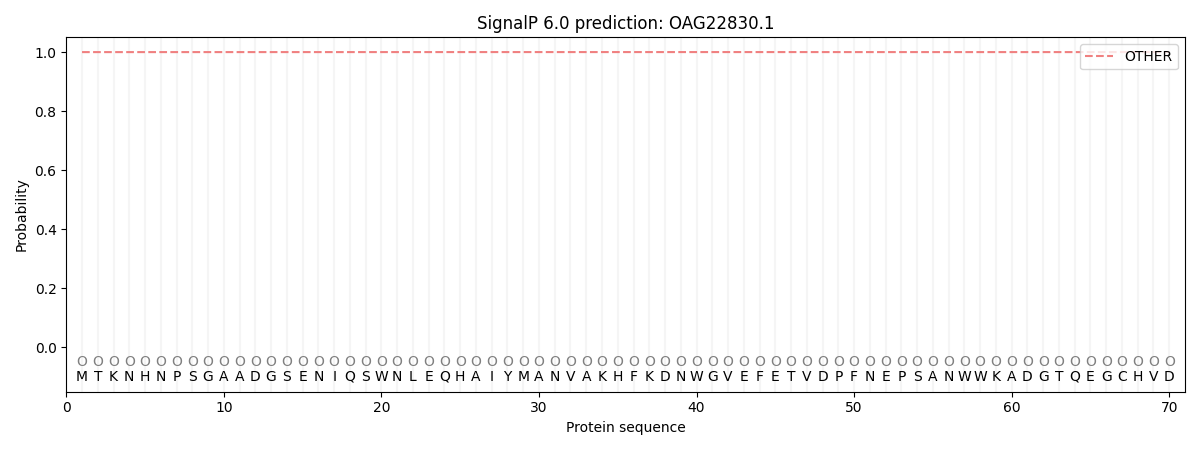
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
1.000050
0.000000
There is no transmembrane helices in OAG22830.1.