You are browsing environment: FUNGIDB
OAG21468.1
Basic Information
help
Species
Alternaria alternata
Lineage
Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata
CAZyme ID
OAG21468.1
CAZy Family
GH31
CAZyme Description
glycoside hydrolase family 79 protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
537
KV441476|CGC5 57733.84
4.3652
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_AalternataSRC1lrK2f
13577
N/A
111
13466
Gene Location
No EC number prediction in OAG21468.1.
Family
Start
End
Evalue
family coverage
GH79
45
404
2.1e-39
0.7472527472527473
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
407104
Glyco_hydro_79C
8.50e-22
427
526
1
103
Glycosyl hydrolase family 79 C-terminal beta domain. This domain is found at the C-terminus of glycosyl hydrolase family 79 proteins. It's function is not yet known.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
5.70e-307
1
537
1
539
5.77e-228
30
537
27
542
5.60e-227
2
537
3
537
1.52e-179
36
530
28
519
4.33e-179
36
530
28
519
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
4.13e-07
83
374
60
325
Crystallographic structure of a bacterial heparanase [Burkholderia pseudomallei],5BWI_B Crystallographic structure of a bacterial heparanase [Burkholderia pseudomallei]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
2.21e-39
12
527
15
540
Beta-glucuronidase OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=An02g11890 PE=1 SV=1
more
2.87e-20
30
447
22
439
Beta-glucuronidase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU00937 PE=1 SV=1
more
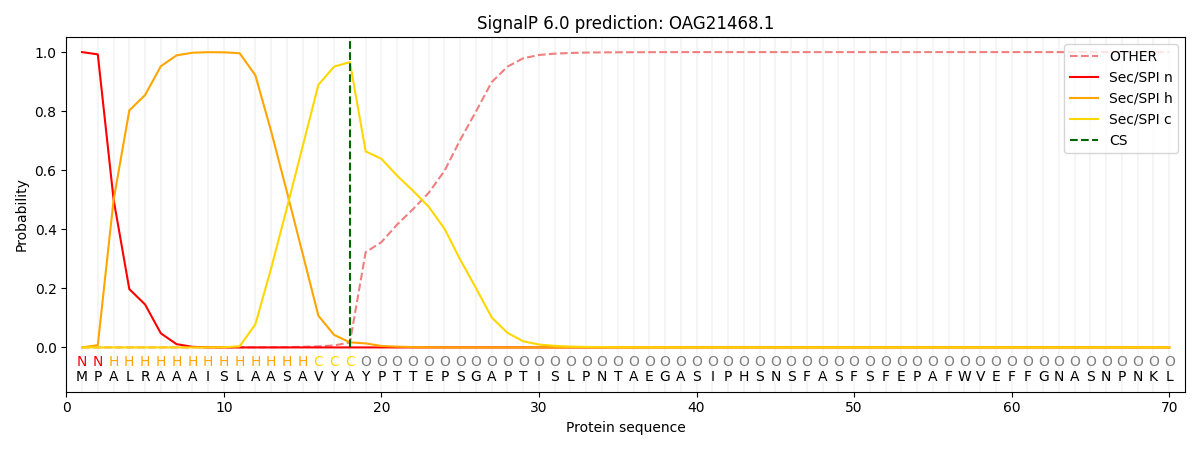
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000584
0.999401
CS pos: 18-19. Pr: 0.9661
There is no transmembrane helices in OAG21468.1.