You are browsing environment: FUNGIDB
CAZyme Information: OAG18459.1
You are here: Home > Sequence: OAG18459.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG18459.1 | |||||||||||
| CAZy Family | GH12 | |||||||||||
| CAZyme Description | FAD/NAD(P)-binding domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 22 | 554 | 5.1e-184 | 0.9817518248175182 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 5.75e-50 | 24 | 553 | 2 | 535 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 398739 | GMC_oxred_C | 2.41e-21 | 423 | 547 | 2 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 235000 | PRK02106 | 4.94e-20 | 29 | 552 | 5 | 532 | choline dehydrogenase; Validated |
| 366272 | GMC_oxred_N | 2.40e-17 | 114 | 303 | 23 | 206 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 224154 | COG1233 | 4.05e-08 | 28 | 73 | 2 | 43 | Phytoene dehydrogenase-related protein [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 552 | 1 | 567 | |
| 2.28e-293 | 8 | 554 | 7 | 549 | |
| 4.77e-293 | 10 | 556 | 10 | 552 | |
| 2.54e-212 | 11 | 554 | 11 | 554 | |
| 2.54e-212 | 11 | 554 | 11 | 554 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.75e-91 | 15 | 553 | 215 | 765 | Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A],4QI7_B Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A] |
|
| 2.77e-89 | 27 | 554 | 5 | 545 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides],4QI5_A Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 2.68e-87 | 17 | 554 | 217 | 767 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 2.20e-83 | 24 | 554 | 2 | 542 | Chain A, cellobiose dehydrogenase [Phanerodontia chrysosporium],1KDG_B Chain B, cellobiose dehydrogenase [Phanerodontia chrysosporium] |
|
| 3.81e-83 | 30 | 554 | 3 | 537 | Chain A, Cellobiose dehydrogenase [Phanerodontia chrysosporium],1NAA_B Chain B, Cellobiose dehydrogenase [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.14e-81 | 12 | 554 | 217 | 769 | Cellobiose dehydrogenase OS=Phanerodontia chrysosporium OX=2822231 GN=CDH-1 PE=1 SV=1 |
|
| 2.15e-16 | 29 | 551 | 4 | 533 | Oxygen-dependent choline dehydrogenase OS=Staphylococcus xylosus OX=1288 GN=betA PE=3 SV=1 |
|
| 6.67e-16 | 29 | 551 | 4 | 533 | Oxygen-dependent choline dehydrogenase OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=betA PE=3 SV=1 |
|
| 9.64e-16 | 18 | 308 | 32 | 324 | Choline dehydrogenase, mitochondrial OS=Rattus norvegicus OX=10116 GN=Chdh PE=1 SV=1 |
|
| 2.22e-15 | 29 | 308 | 40 | 319 | Choline dehydrogenase, mitochondrial OS=Homo sapiens OX=9606 GN=CHDH PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
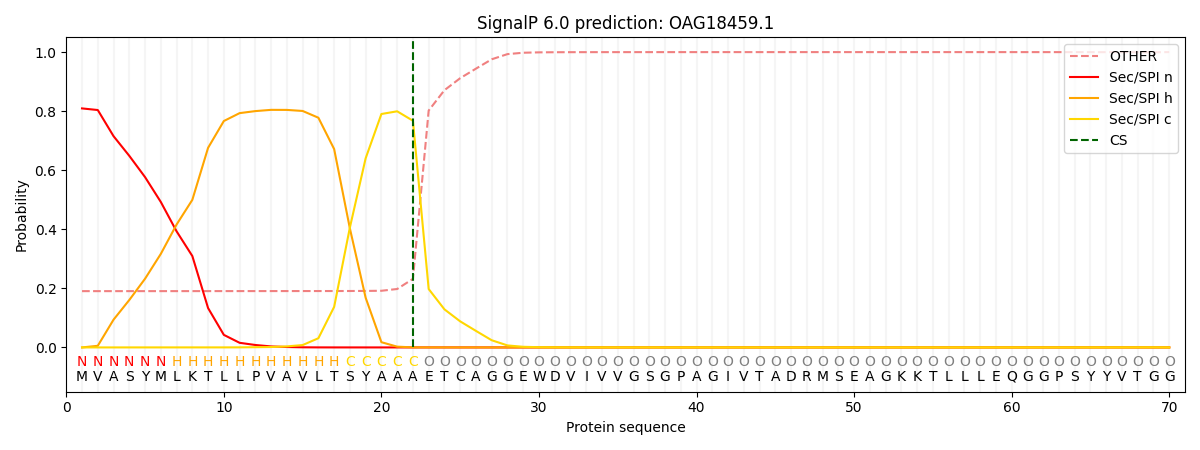
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.197966 | 0.802001 | CS pos: 22-23. Pr: 0.7676 |
