You are browsing environment: FUNGIDB
CAZyme Information: OAG18428.1
You are here: Home > Sequence: OAG18428.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG18428.1 | |||||||||||
| CAZy Family | GH12 | |||||||||||
| CAZyme Description | endo-1,4-beta-xylanase B precursor | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 43 | 210 | 4e-54 | 0.943502824858757 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 1.62e-78 | 41 | 213 | 3 | 175 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.11e-77 | 1 | 220 | 1 | 221 | |
| 2.06e-73 | 1 | 220 | 1 | 223 | |
| 1.68e-72 | 1 | 220 | 1 | 223 | |
| 2.16e-70 | 1 | 218 | 1 | 220 | |
| 3.41e-69 | 1 | 218 | 1 | 219 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.05e-31 | 41 | 214 | 4 | 174 | Chain A, Endo-1,4-beta-xylanase I [Trichoderma reesei] |
|
| 7.46e-16 | 61 | 218 | 34 | 189 | Chain A, Endo-1,4-beta-xylanase 2 [Trichoderma reesei],4XQ4_A Chain A, Endo-1,4-beta-xylanase 2 [Trichoderma reesei],4XQ4_B Chain B, Endo-1,4-beta-xylanase 2 [Trichoderma reesei],6JXL_A Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose [Trichoderma reesei RUT C-30],6K9O_A Crystal Structure Analysis of Protein [Trichoderma reesei RUT C-30],6KWD_A Crystal Structure Analysis of Endo-beta-1,4-Xylanase II Complexed with Xylotriose [Trichoderma reesei RUT C-30] |
|
| 7.60e-16 | 42 | 218 | 13 | 190 | HIGH-RESOLUTION STRUCTURES OF XYLANASES FROM B. CIRCULANS AND T. HARZIANUM IDENTIFY A NEW FOLDING PATTERN AND IMPLICATIONS FOR THE ATOMIC BASIS OF THE CATALYSIS [Trichoderma harzianum] |
|
| 7.60e-16 | 61 | 218 | 35 | 190 | Chain A, Endo-1,4-beta-xylanase 2 [Trichoderma reesei RUT C-30],6KW9_A Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose [Trichoderma reesei RUT C-30],6KWF_A Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose [Trichoderma reesei RUT C-30] |
|
| 1.19e-15 | 31 | 220 | 3 | 193 | Chain A, endoxylanase 11A [Thermochaetoides thermophila],1XNK_B Chain B, endoxylanase 11A [Thermochaetoides thermophila] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.81e-34 | 1 | 214 | 1 | 225 | Endo-1,4-beta-xylanase 1 OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=xyn1 PE=1 SV=1 |
|
| 3.81e-34 | 1 | 214 | 1 | 225 | Endo-1,4-beta-xylanase 1 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=TRIREDRAFT_74223 PE=1 SV=1 |
|
| 3.98e-24 | 50 | 217 | 59 | 224 | Endo-1,4-beta-xylanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xlnB PE=3 SV=2 |
|
| 5.95e-24 | 1 | 217 | 1 | 227 | Probable endo-1,4-beta-xylanase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xlnA PE=3 SV=1 |
|
| 6.48e-24 | 1 | 217 | 1 | 231 | Probable endo-1,4-beta-xylanase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=xlnA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
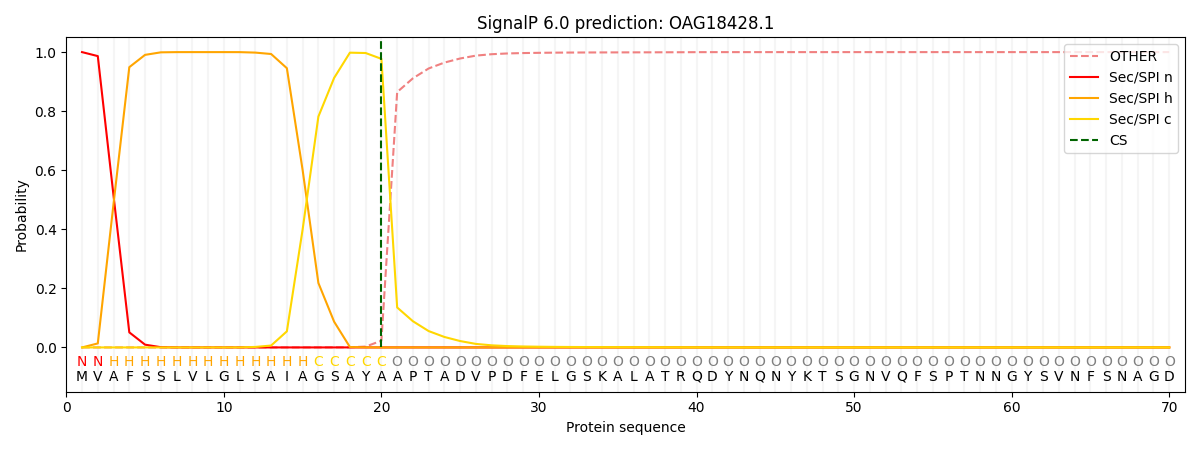
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000258 | 0.999702 | CS pos: 20-21. Pr: 0.9772 |
