You are browsing environment: FUNGIDB
CAZyme Information: OAG17278.1
You are here: Home > Sequence: OAG17278.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG17278.1 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | Phosphomevalonate kinase [Source:UniProtKB/TrEMBL;Acc:A0A177DCK2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 306 | 541 | 9e-47 | 0.462882096069869 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235028 | PRK02304 | 1.57e-31 | 958 | 1149 | 1 | 175 | adenine phosphoribosyltransferase; Provisional |
| 223577 | Apt | 1.38e-27 | 961 | 1147 | 6 | 177 | Adenine/guanine phosphoribosyltransferase or related PRPP-binding protein [Nucleotide transport and metabolism]. |
| 177930 | PLN02293 | 3.34e-25 | 960 | 1134 | 14 | 176 | adenine phosphoribosyltransferase |
| 398111 | P-mevalo_kinase | 1.55e-22 | 768 | 894 | 1 | 111 | Phosphomevalonate kinase. Phosphomevalonate kinase (EC:2.7.4.2) catalyzes the phosphorylation of 5-phosphomevalonate into 5-diphosphomevalonate, an essential step in isoprenoid biosynthesis via the mevalonate pathway. This family represents the animal type of the enzyme. The other is the ERG8 type, found in plants and fungi, and some bacteria (see pfam00288). |
| 223354 | GlcD | 4.27e-19 | 286 | 522 | 3 | 225 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.02e-13 | 314 | 749 | 62 | 483 | |
| 1.58e-12 | 318 | 749 | 64 | 475 | |
| 2.57e-12 | 314 | 530 | 120 | 316 | |
| 4.49e-12 | 314 | 535 | 119 | 321 | |
| 3.55e-11 | 316 | 507 | 71 | 241 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.48e-17 | 971 | 1143 | 18 | 182 | Crystal Structure Of Adenine Phosphoribosyltransferase [Saccharomyces cerevisiae],1G2Q_A Crystal Structure Of Adenine Phosphoribosyltransferase [Saccharomyces cerevisiae],1G2Q_B Crystal Structure Of Adenine Phosphoribosyltransferase [Saccharomyces cerevisiae],5VJN_A Crystal Structure of Adenine Phosphoribosyltransferase from Saccharomyces cerevisiae Complexed with D-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (D-DIAB) and Adenine [Saccharomyces cerevisiae],5VJN_B Crystal Structure of Adenine Phosphoribosyltransferase from Saccharomyces cerevisiae Complexed with D-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (D-DIAB) and Adenine [Saccharomyces cerevisiae],5VJP_A Crystal Structure of Adenine Phosphoribosyltransferase from Saccharomyces cerevisiae Complexed with L-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (L-DIAB) and Adenine [Saccharomyces cerevisiae],5VJP_B Crystal Structure of Adenine Phosphoribosyltransferase from Saccharomyces cerevisiae Complexed with L-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (L-DIAB) and Adenine [Saccharomyces cerevisiae] |
|
| 9.63e-17 | 313 | 512 | 37 | 214 | Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 3 (P1) [Paenarthrobacter nicotinovorans],2BVF_B Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 3 (P1) [Paenarthrobacter nicotinovorans],2BVG_A Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 1 (P21) [Paenarthrobacter nicotinovorans],2BVG_B Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 1 (P21) [Paenarthrobacter nicotinovorans],2BVG_C Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 1 (P21) [Paenarthrobacter nicotinovorans],2BVG_D Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 1 (P21) [Paenarthrobacter nicotinovorans],2BVH_A Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 2 (P21) [Paenarthrobacter nicotinovorans],2BVH_B Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 2 (P21) [Paenarthrobacter nicotinovorans],2BVH_C Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 2 (P21) [Paenarthrobacter nicotinovorans],2BVH_D Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 2 (P21) [Paenarthrobacter nicotinovorans] |
|
| 3.91e-16 | 959 | 1131 | 6 | 168 | Crystal structure of Adenine phosphoribosyltransferase from Francisella tularensis. [Francisella tularensis],5YW2_B Crystal structure of Adenine phosphoribosyltransferase from Francisella tularensis. [Francisella tularensis],5YW2_C Crystal structure of Adenine phosphoribosyltransferase from Francisella tularensis. [Francisella tularensis],5YW2_D Crystal structure of Adenine phosphoribosyltransferase from Francisella tularensis. [Francisella tularensis],5YW5_A Crystal structure of Adenine phosphoribosyltransferase from Francisella tularensis in complex with adenine [Francisella tularensis],5YW5_B Crystal structure of Adenine phosphoribosyltransferase from Francisella tularensis in complex with adenine [Francisella tularensis],5YW5_C Crystal structure of Adenine phosphoribosyltransferase from Francisella tularensis in complex with adenine [Francisella tularensis],5YW5_D Crystal structure of Adenine phosphoribosyltransferase from Francisella tularensis in complex with adenine [Francisella tularensis] |
|
| 4.00e-16 | 958 | 1142 | 4 | 180 | Crystal Structure of F173G Mutant of Human APRT [Homo sapiens],4X45_B Crystal Structure of F173G Mutant of Human APRT [Homo sapiens] |
|
| 1.78e-15 | 958 | 1131 | 2 | 166 | Crystal Structure of Human APRT wild type in complex with PRPP and Mg2+ [Homo sapiens],6FCH_B Crystal Structure of Human APRT wild type in complex with PRPP and Mg2+ [Homo sapiens],6FCL_A Crystal Structure of Human APRT wild type in complex with AMP [Homo sapiens],6FCL_B Crystal Structure of Human APRT wild type in complex with AMP [Homo sapiens],6HGP_A Crystal Structure of Human APRT wild type in complex with Phosphate ion. [Homo sapiens],6HGP_B Crystal Structure of Human APRT wild type in complex with Phosphate ion. [Homo sapiens],6HGR_A Crystal Structure of Human APRT wild type in complex with IMP [Homo sapiens],6HGR_B Crystal Structure of Human APRT wild type in complex with IMP [Homo sapiens],6HGS_A Crystal Structure of Human APRT wild type in complex with GMP [Homo sapiens],6HGS_B Crystal Structure of Human APRT wild type in complex with GMP [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.96e-23 | 318 | 746 | 51 | 437 | FAD-linked oxidoreductase DDB_G0289697 OS=Dictyostelium discoideum OX=44689 GN=DDB_G0289697 PE=2 SV=1 |
|
| 3.61e-21 | 326 | 757 | 44 | 443 | Uncharacterized FAD-linked oxidoreductase YgaK OS=Bacillus subtilis (strain 168) OX=224308 GN=ygaK PE=3 SV=4 |
|
| 1.92e-19 | 960 | 1149 | 3 | 177 | Adenine phosphoribosyltransferase OS=Erythrobacter litoralis (strain HTCC2594) OX=314225 GN=apt PE=3 SV=1 |
|
| 3.73e-19 | 958 | 1147 | 4 | 179 | Adenine phosphoribosyltransferase OS=Cricetulus griseus OX=10029 GN=APRT PE=3 SV=2 |
|
| 9.56e-19 | 959 | 1098 | 18 | 147 | Adenine phosphoribosyltransferase OS=Bifidobacterium longum subsp. infantis (strain ATCC 15697 / DSM 20088 / JCM 1222 / NCTC 11817 / S12) OX=391904 GN=apt PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
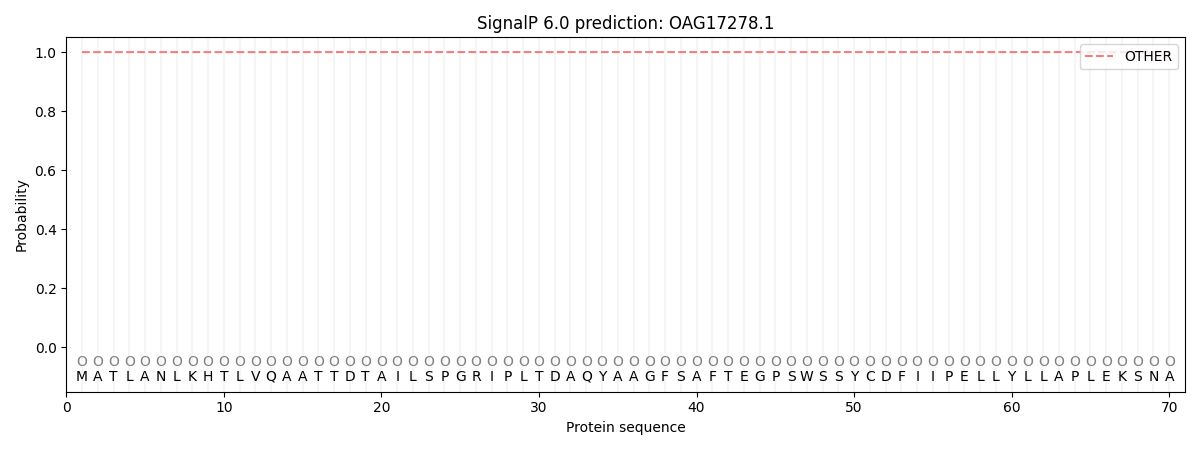
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000030 | 0.000021 |
