You are browsing environment: FUNGIDB
CAZyme Information: OAG17102.1
You are here: Home > Sequence: OAG17102.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG17102.1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | glycoside hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.58:29 | 3.2.1.21:6 | 3.2.1.75:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 82 | 375 | 1e-107 | 0.9966996699669967 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 3.85e-64 | 35 | 405 | 17 | 395 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 8.68e-07 | 68 | 207 | 1 | 135 | Cellulase (glycosyl hydrolase family 5). |
| 396834 | Glyco_hydro_42 | 0.003 | 82 | 143 | 5 | 63 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.78e-282 | 1 | 404 | 1 | 405 | |
| 6.13e-263 | 1 | 405 | 1 | 407 | |
| 1.53e-261 | 1 | 405 | 241 | 650 | |
| 5.70e-191 | 7 | 405 | 19 | 416 | |
| 6.52e-176 | 8 | 397 | 8 | 413 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.48e-130 | 29 | 405 | 2 | 392 | Exo-b-(1,3)-glucanase From Candida Albicans [Candida albicans] |
|
| 2.98e-130 | 29 | 405 | 2 | 392 | Exo-b-(1,3)-glucanase From Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
|
| 3.64e-130 | 29 | 405 | 8 | 398 | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A [Candida albicans] |
|
| 1.03e-129 | 29 | 405 | 8 | 398 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
|
| 1.03e-129 | 29 | 405 | 8 | 398 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.11e-157 | 9 | 405 | 10 | 417 | Glucan 1,3-beta-glucosidase OS=Blumeria graminis OX=34373 PE=3 SV=1 |
|
| 1.07e-147 | 13 | 404 | 16 | 415 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=exgA PE=3 SV=1 |
|
| 8.50e-142 | 19 | 404 | 14 | 405 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=exgA PE=3 SV=1 |
|
| 8.50e-142 | 19 | 404 | 14 | 405 | Glucan 1,3-beta-glucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=exgA PE=1 SV=1 |
|
| 4.98e-141 | 11 | 404 | 17 | 415 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
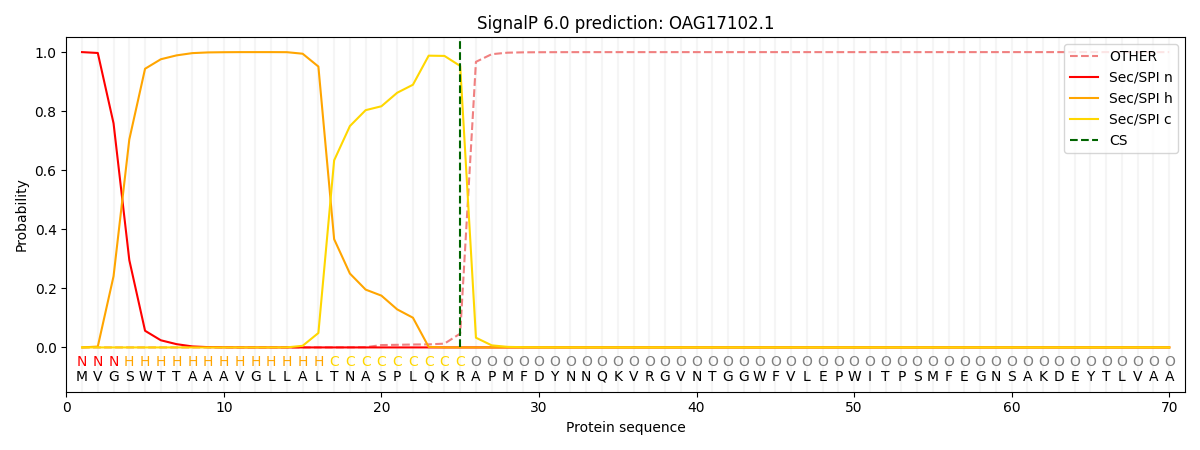
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000388 | 0.999566 | CS pos: 25-26. Pr: 0.9531 |
