You are browsing environment: FUNGIDB
CAZyme Information: OAG16706.1
You are here: Home > Sequence: OAG16706.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
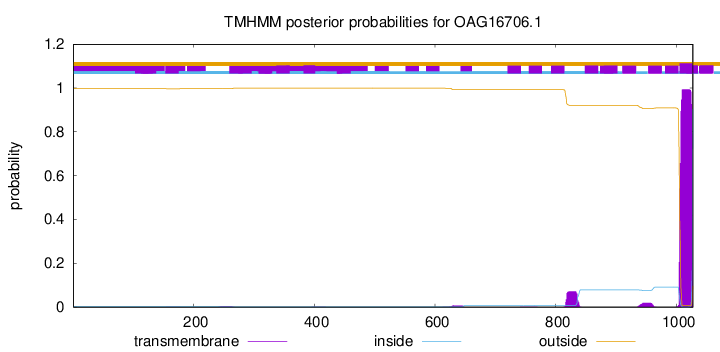
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG16706.1 | |||||||||||
| CAZy Family | CE12 | |||||||||||
| CAZyme Description | 3-methylcrotonyl-CoA carboxylase 2 [Source:UniProtKB/TrEMBL;Acc:A0A177DB44] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 709773; End:714399 Strand: + | |||||||||||
Full Sequence Download help
| MVPRAPSREA LTLLKHSVRP SVVVRSQTQR CLLSSTTRPT LPSHAQSQIP QKSIRNTATF | 60 |
| THSHHAEAVS VIPTTVDTSN ADFKENKRQM DEATENLVNL HTKIAQGGPQ KARDKHIQRG | 120 |
| KMLVRDRISA LIDPGTPFLE LSQMAGYDMY GEDDVPAGGI VTGIGTVNGV QCMVVANDAT | 180 |
| VKGGTYYPVT VKKHLRAQTI AQENRLPCIY LVDSGGANLP NQADVFPDVN HFGRIFYNQA | 240 |
| RMSAMGIPQI SVVMGPCTAG GAYVPSMSDE NIIVENQGHI FLAGPPLVKA ATGEVVSAED | 300 |
| LGGGKLHSEV SGVTDYLAVD DAHALVLARR SVGNLNWHRN QTVTQQPAFQ EPLYDPQELS | 360 |
| GIVGTNLRRQ IPIHEIIARI VDGSSFDEFK PLYGSTLVTG FAKIYGHPVG IVANNGILFS | 420 |
| ESSLKGAHFV QLCGKRHIPL IFLQNISGFM VGQDAEKGGI AKNGAKLVTA VSCVDVPKFT | 480 |
| VVVGSSAGAG NYGMCGRAYS PRLLFTWPNA KTSVMGAEQL SSVMEAVGKK VDPALKARIE | 540 |
| HESEATFGSA RLWDDGIIPP EHTRRVLGMG LQMACGGQNK GVEKESTWAS IKDVSSQVAK | 600 |
| QLVAQYATID DKGIHVLGGY PGILYPPYYW WQAGAMFGTL LDYWHYTGDD QYNEMVREGL | 660 |
| IHQFGEHQDL MPSNQSKNEG NDDQVFWAFS MIAAAEYKLK DPPSDQPGWL AMTQSVFNQF | 720 |
| VGRYQKEVSD GTCGGGMRWQ IYPWLNGWTY KNTASNGGLF HLGARLAMYT KNDTYARWAE | 780 |
| KAFDWMAQSP ILPGDGQVYD GTSITTNPPC HDADQTPWTY NYGIMIAGAA YMYNYTNGAE | 840 |
| KWRTELTKFI NKVAIFFPEE KGGVMVEICE ANQVCDADQE SFKAYLARWL GITMQMAPEF | 900 |
| TPQILPKLQK SAVAAAQTCE GPSQHNGGPH QCGMKWYATG FDGIGGVGPQ MTALNVISVL | 960 |
| NAQRVPPPYS RDTGGTSEGN PGLGSGNDDD RLPKFQSEIT TGDKAGAAIL TILMFGLFAG | 1020 |
| GAVWILM | 1027 |
Enzyme Prediction help
| EC | 3.2.1.101:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 584 | 964 | 5.7e-97 | 0.946927374301676 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178415 | PLN02820 | 0.0 | 38 | 574 | 7 | 554 | 3-methylcrotonyl-CoA carboxylase, beta chain |
| 227136 | MmdA | 0.0 | 81 | 583 | 2 | 514 | Acetyl-CoA carboxylase, carboxyltransferase component [Lipid transport and metabolism]. |
| 395825 | Carboxyl_trans | 1.07e-165 | 115 | 574 | 1 | 476 | Carboxyl transferase domain. All of the members in this family are biotin dependent carboxylases. The carboxyl transferase domain carries out the following reaction; transcarboxylation from biotin to an acceptor molecule. There are two recognized types of carboxyl transferase. One of them uses acyl-CoA and the other uses 2-oxoacid as the acceptor molecule of carbon dioxide. All of the members in this family utilize acyl-CoA as the acceptor molecule. |
| 397638 | Glyco_hydro_76 | 3.25e-157 | 590 | 954 | 1 | 348 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 130187 | mmdA | 1.66e-80 | 92 | 580 | 1 | 501 | methylmalonyl-CoA decarboxylase alpha subunit. This model describes methymalonyl-CoA decarboxylase aplha subunit in archaea and bacteria. Metylmalonyl-CoA decarboxylase Na+ pump is a representative of a class of Na+ transport decarboxylases that couples the energy derived by decarboxylation of carboxylic acid substrates to drive the extrusion of Na+ ion across the membrane. [Energy metabolism, ATP-proton motive force interconversion, Energy metabolism, Fermentation, Transport and binding proteins, Cations and iron carrying compounds] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAE6998605.1|GH76 | 2.03e-289 | 589 | 1026 | 39 | 478 |
| CBY02446.1|GH76 | 1.13e-276 | 586 | 1026 | 40 | 480 |
| QRC95121.1|GH76 | 3.96e-241 | 590 | 1026 | 53 | 491 |
| QSZ36627.1|GH76 | 1.19e-121 | 589 | 1026 | 33 | 451 |
| APA11655.1|GH76 | 3.28e-121 | 590 | 1026 | 33 | 450 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3U9R_B | 1.48e-206 | 69 | 574 | 21 | 540 | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC), beta subunit [Pseudomonas aeruginosa],3U9S_B Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex [Pseudomonas aeruginosa],3U9S_D Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex [Pseudomonas aeruginosa],3U9S_F Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex [Pseudomonas aeruginosa],3U9S_H Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex [Pseudomonas aeruginosa],3U9S_J Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex [Pseudomonas aeruginosa],3U9S_L Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex [Pseudomonas aeruginosa],3U9T_B Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, free enzyme [Pseudomonas aeruginosa] |
| 4Q0G_A | 1.34e-196 | 91 | 573 | 14 | 513 | Chain A, Probable acetyl-/propionyl-CoA carboxylase (Beta subunit) AccD1 [Mycobacterium tuberculosis],4Q0G_B Chain B, Probable acetyl-/propionyl-CoA carboxylase (Beta subunit) AccD1 [Mycobacterium tuberculosis],4Q0G_C Chain C, Probable acetyl-/propionyl-CoA carboxylase (Beta subunit) AccD1 [Mycobacterium tuberculosis] |
| 5IKL_B | 2.15e-142 | 72 | 586 | 3 | 531 | Crystal structure of P. aeruginosa geranyl-CoA carboxylase (GCC), beta subunit [Pseudomonas aeruginosa PAO1],5IKL_D Crystal structure of P. aeruginosa geranyl-CoA carboxylase (GCC), beta subunit [Pseudomonas aeruginosa PAO1],5IKL_F Crystal structure of P. aeruginosa geranyl-CoA carboxylase (GCC), beta subunit [Pseudomonas aeruginosa PAO1] |
| 6RY0_A | 3.59e-88 | 618 | 984 | 57 | 425 | Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY1_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY2_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY5_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY6_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY7_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495] |
| 1ON3_A | 9.81e-73 | 90 | 573 | 12 | 505 | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) [Propionibacterium freudenreichii],1ON3_B Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) [Propionibacterium freudenreichii],1ON3_C Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) [Propionibacterium freudenreichii],1ON3_D Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) [Propionibacterium freudenreichii],1ON3_E Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) [Propionibacterium freudenreichii],1ON3_F Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) [Propionibacterium freudenreichii],1ON9_A Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with hydrolyzed methylmalonyl-coenzyme a bound) [Propionibacterium freudenreichii],1ON9_B Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with hydrolyzed methylmalonyl-coenzyme a bound) [Propionibacterium freudenreichii],1ON9_C Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with hydrolyzed methylmalonyl-coenzyme a bound) [Propionibacterium freudenreichii],1ON9_D Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with hydrolyzed methylmalonyl-coenzyme a bound) [Propionibacterium freudenreichii],1ON9_E Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with hydrolyzed methylmalonyl-coenzyme a bound) [Propionibacterium freudenreichii],1ON9_F Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with hydrolyzed methylmalonyl-coenzyme a bound) [Propionibacterium freudenreichii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q9V9A7|MCCB_DROME | 1.23e-225 | 65 | 574 | 23 | 563 | Probable methylcrotonoyl-CoA carboxylase beta chain, mitochondrial OS=Drosophila melanogaster OX=7227 GN=Mccc2 PE=2 SV=1 |
| sp|Q5XIT9|MCCB_RAT | 6.57e-224 | 64 | 574 | 23 | 548 | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial OS=Rattus norvegicus OX=10116 GN=Mccc2 PE=2 SV=1 |
| sp|Q3ULD5|MCCB_MOUSE | 1.86e-223 | 64 | 574 | 23 | 548 | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial OS=Mus musculus OX=10090 GN=Mccc2 PE=1 SV=1 |
| sp|Q9HCC0|MCCB_HUMAN | 1.38e-214 | 64 | 584 | 23 | 555 | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial OS=Homo sapiens OX=9606 GN=MCCC2 PE=1 SV=1 |
| sp|P34385|MCCB_CAEEL | 4.74e-211 | 45 | 572 | 43 | 591 | Probable methylcrotonoyl-CoA carboxylase beta chain, mitochondrial OS=Caenorhabditis elegans OX=6239 GN=F02A9.10 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
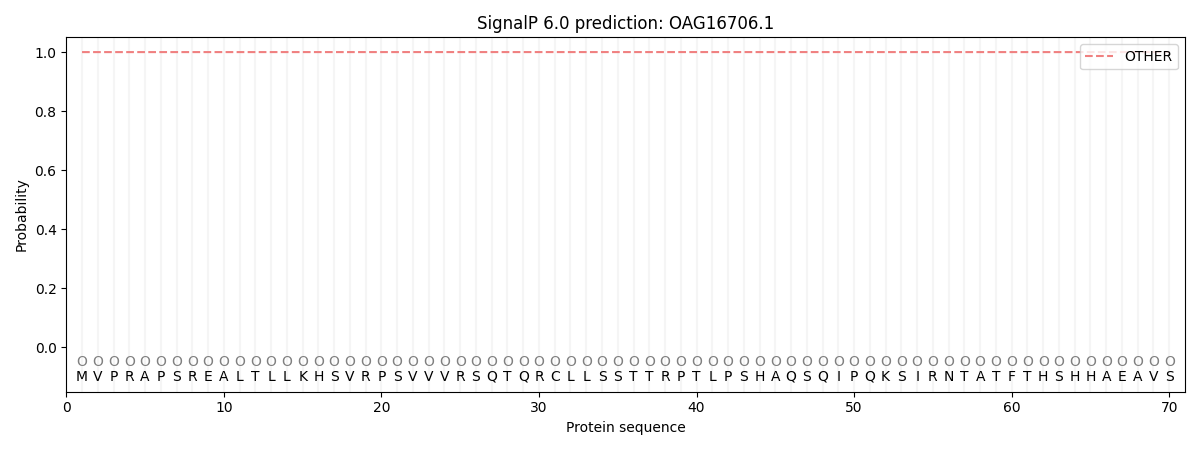
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000057 | 0.000000 |