You are browsing environment: FUNGIDB
CAZyme Information: OAG15534.1
You are here: Home > Sequence: OAG15534.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG15534.1 | |||||||||||
| CAZy Family | AA8|AA3 | |||||||||||
| CAZyme Description | alpha/beta-hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 45 | 176 | 2.8e-18 | 0.6035242290748899 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226040 | LpqC | 1.75e-14 | 12 | 278 | 13 | 306 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.45e-102 | 22 | 279 | 90 | 355 | |
| 1.50e-102 | 22 | 279 | 39 | 304 | |
| 8.55e-100 | 23 | 279 | 37 | 300 | |
| 1.80e-99 | 16 | 279 | 34 | 298 | |
| 2.16e-99 | 16 | 279 | 35 | 299 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.27e-87 | 17 | 277 | 15 | 268 | Probable feruloyl esterase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=faeC PE=3 SV=1 |
|
| 3.34e-86 | 13 | 276 | 11 | 267 | Probable feruloyl esterase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=faeC PE=3 SV=1 |
|
| 3.34e-86 | 13 | 276 | 11 | 267 | Feruloyl esterase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=faeC PE=1 SV=1 |
|
| 4.50e-86 | 13 | 279 | 13 | 289 | Feruloyl esterase D OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=faeD-3.544 PE=1 SV=1 |
|
| 2.33e-84 | 22 | 276 | 22 | 269 | Probable feruloyl esterase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=faeC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
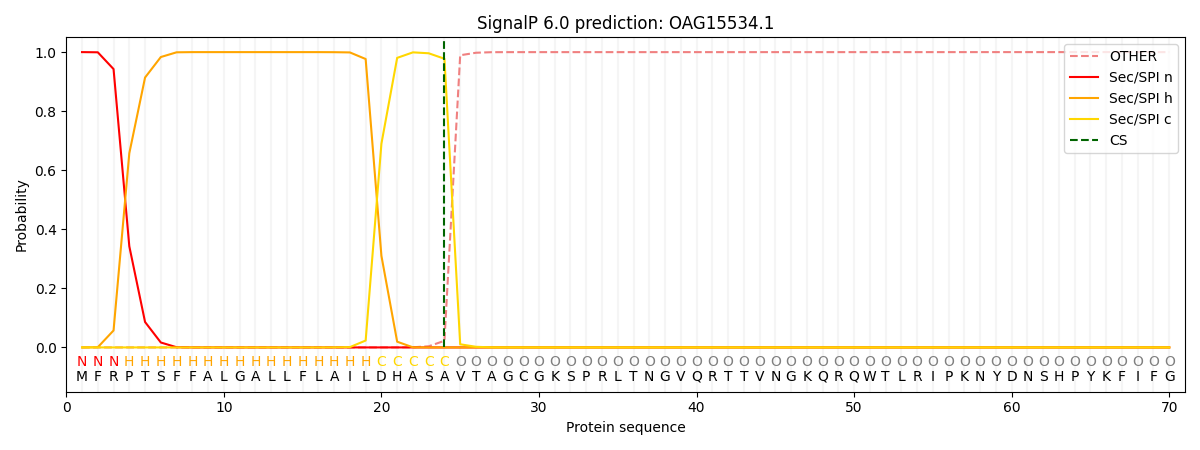
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000202 | 0.999739 | CS pos: 24-25. Pr: 0.9775 |
