You are browsing environment: FUNGIDB
CAZyme Information: OAG15446.1
You are here: Home > Sequence: OAG15446.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG15446.1 | |||||||||||
| CAZy Family | AA8 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 76 | 257 | 4.3e-20 | 0.5901060070671378 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398399 | Dickkopf_N | 0.002 | 588 | 636 | 1 | 49 | Dickkopf N-terminal cysteine-rich region. Dickkopf proteins are a class of Wnt antagonists. They possess two conserved cysteine-rich regions. This family represents the N-terminal one. The C-terminal region has been found to share significant sequence similarity to the colipase fold, pfam01114, pfam02740. |
| 411343 | exchanger_TraA | 0.008 | 578 | 628 | 356 | 403 | outer membrane exchange protein TraA. TraA, together with its partner TraB, mediates a large scale exchange of outer membrane lipoproteins, and lipids, between closely related strains or clonally identical cells, certain delta-proteobacterial species such as Myxococcus xanthus. The exchange mechanism is likely to involve fusion of outer membrane, probably done to coordinate the social behaviors these bacteria display. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.93e-293 | 1 | 671 | 1 | 610 | |
| 4.59e-240 | 1 | 457 | 2 | 461 | |
| 2.97e-32 | 17 | 335 | 8 | 317 | |
| 2.05e-31 | 32 | 345 | 31 | 332 | |
| 1.00e-22 | 22 | 344 | 11 | 314 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
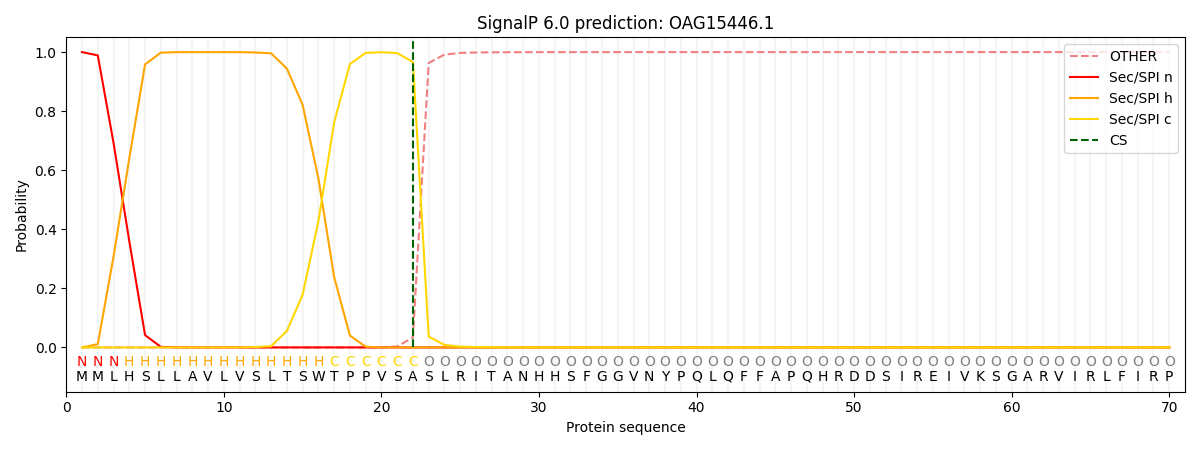
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000391 | 0.999575 | CS pos: 22-23. Pr: 0.9662 |
