You are browsing environment: FUNGIDB
CAZyme Information: OAG15368.1
Basic Information
help
| Species |
Alternaria alternata
|
| Lineage |
Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata
|
| CAZyme ID |
OAG15368.1
|
| CAZy Family |
AA7 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 689 |
|
75871.04 |
4.9355 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AalternataSRC1lrK2f |
13577 |
N/A |
111 |
13466
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| CE18 |
249 |
616 |
6.6e-194 |
0.9945504087193461 |
| CBM87 |
40 |
248 |
7e-89 |
0.9954128440366973 |
OAG15368.1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
7 |
689 |
5 |
687 |
| 0.0 |
1 |
689 |
1 |
693 |
| 0.0 |
2 |
689 |
4 |
699 |
| 9.09e-253 |
24 |
687 |
34 |
706 |
| 4.34e-246 |
32 |
689 |
29 |
699 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6.24e-243 |
33 |
689 |
23 |
684 |
Crystal structure of Agd3 a novel carbohydrate deacetylase [Aspergillus fumigatus Af293] |
OAG15368.1 has no Swissprot hit.
This protein is predicted as SP
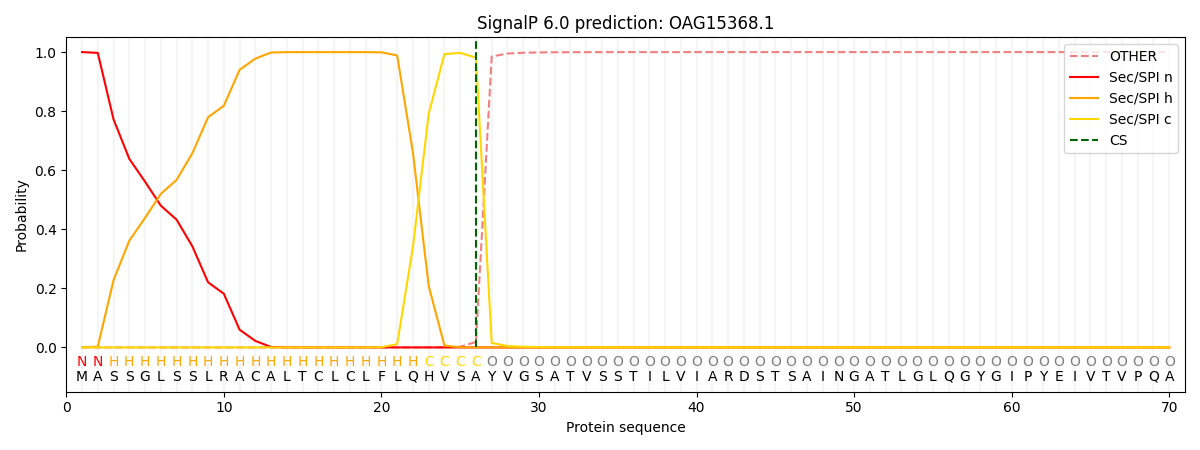
| Other |
SP_Sec_SPI |
CS Position |
| 0.000214 |
0.999759 |
CS pos: 26-27. Pr: 0.9813 |
There is no transmembrane helices in OAG15368.1.

