You are browsing environment: FUNGIDB
CAZyme Information: OAG14841.1
You are here: Home > Sequence: OAG14841.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG14841.1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | GFO_IDH_MocA domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A177D686] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 8 | 157 | 2.1e-24 | 0.38847117794486213 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 99740 | AHBA_syn | 2.96e-122 | 409 | 756 | 3 | 352 | 3-amino-5-hydroxybenzoic acid synthase family (AHBA_syn). AHBA_syn family belongs to pyridoxal phosphate (PLP)-dependent aspartate aminotransferase superfamily (fold I). The members of this CD are involved in various biosynthetic pathways for secondary metabolites. Some well studied proteins in this CD are AHBA_synthase, protein product of pleiotropic regulatory gene degT, Arnb aminotransferase and pilin glycosylation protein. The prototype of this family, the AHBA_synthase, is a dimeric PLP dependent enzyme. AHBA_syn is the terminal enzyme of 3-amino-5-hydroxybenzoic acid (AHBA) formation which is involved in the biosynthesis of ansamycin antibiotics, including rifamycin B. Some members of this CD are involved in 4-amino-6-deoxy-monosaccharide D-perosamine synthesis. Perosamine is an important element in the glycosylation of several cell products, such as antibiotics and lipopolysaccharides of gram-positive and gram-negative bacteria. The pilin glycosylation protein encoded by gene pglA, is a galactosyltransferase involved in pilin glycosylation. Additionally, this CD consists of ArnB (PmrH) aminotransferase, a 4-amino-4-deoxy-L-arabinose lipopolysaccharide-modifying enzyme. This CD also consists of several predicted pyridoxal phosphate-dependent enzymes apparently involved in regulation of cell wall biogenesis. The catalytic lysine which is present in all characterized PLP dependent enzymes is replaced by histidine in some members of this CD. |
| 223476 | WecE | 1.16e-115 | 391 | 760 | 1 | 372 | dTDP-4-amino-4,6-dideoxygalactose transaminase [Cell wall/membrane/envelope biogenesis]. |
| 395827 | DegT_DnrJ_EryC1 | 1.09e-108 | 409 | 756 | 9 | 360 | DegT/DnrJ/EryC1/StrS aminotransferase family. The members of this family are probably all pyridoxal-phosphate-dependent aminotransferase enzymes with a variety of molecular functions. The family includes StsA, StsC and StsS. The aminotransferase activity was demonstrated for purified StsC protein as the L-glutamine:scyllo-inosose aminotransferase EC:2.6.1.50, which catalyzes the first amino transfer in the biosynthesis of the streptidine subunit of streptomycin. |
| 183263 | PRK11658 | 4.10e-50 | 411 | 760 | 20 | 378 | UDP-4-amino-4-deoxy-L-arabinose aminotransferase. |
| 223745 | MviM | 4.11e-50 | 4 | 332 | 1 | 336 | Predicted dehydrogenase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.24e-66 | 389 | 743 | 2 | 350 | |
| 4.48e-29 | 423 | 760 | 36 | 385 | |
| 4.74e-29 | 423 | 739 | 675 | 988 | |
| 5.54e-29 | 411 | 747 | 19 | 357 | |
| 9.33e-27 | 423 | 756 | 656 | 986 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.80e-103 | 383 | 756 | 1 | 375 | Crystal structure of a DegT DnrJ EryC1 StrS aminotransferase from Brucella abortus [Brucella abortus 2308],4QGR_B Crystal structure of a DegT DnrJ EryC1 StrS aminotransferase from Brucella abortus [Brucella abortus 2308] |
|
| 7.42e-94 | 391 | 747 | 2 | 349 | WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with the Cofactor PMP [Pseudomonas aeruginosa],3NU7_B WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with the Cofactor PMP [Pseudomonas aeruginosa],3NUB_A WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with Product as the External Aldimine [Pseudomonas aeruginosa],3NUB_B WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with Product as the External Aldimine [Pseudomonas aeruginosa] |
|
| 5.67e-93 | 391 | 747 | 2 | 349 | WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with the Internal Aldimine [Pseudomonas aeruginosa],3NU8_B WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with the Internal Aldimine [Pseudomonas aeruginosa] |
|
| 7.31e-93 | 391 | 747 | 2 | 349 | X-ray crystal structure of the Wbpe (WlbE) aminotransferase from pseudomonas aeruginosa as the PLP internal aldimine adduct with lysine 185 [Pseudomonas aeruginosa PAO1],3NYU_B X-ray crystal structure of the Wbpe (WlbE) aminotransferase from pseudomonas aeruginosa as the PLP internal aldimine adduct with lysine 185 [Pseudomonas aeruginosa PAO1],3NYU_C X-ray crystal structure of the Wbpe (WlbE) aminotransferase from pseudomonas aeruginosa as the PLP internal aldimine adduct with lysine 185 [Pseudomonas aeruginosa PAO1],3NYU_D X-ray crystal structure of the Wbpe (WlbE) aminotransferase from pseudomonas aeruginosa as the PLP internal aldimine adduct with lysine 185 [Pseudomonas aeruginosa PAO1] |
|
| 7.31e-93 | 391 | 747 | 2 | 349 | X-ray structure of the K185A mutant of WbpE (WlbE) from pseudomonas aeruginosa in complex with PLP at 1.45 angstrom resolution [Pseudomonas aeruginosa PAO1],3NYT_A X-ray crystal structure of the WlbE (WpbE) aminotransferase from pseudomonas aeruginosa, mutation K185A, in complex with the PLP external aldimine adduct with UDP-3-amino-2-N-acetyl-glucuronic acid, at 1.3 angstrom resolution [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.82e-93 | 391 | 747 | 2 | 349 | UDP-2-acetamido-2-deoxy-3-oxo-D-glucuronate aminotransferase OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=wbpE PE=1 SV=1 |
|
| 2.53e-74 | 391 | 757 | 10 | 371 | dTDP-3-amino-3,4,6-trideoxy-alpha-D-glucose transaminase OS=Streptomyces venezuelae OX=54571 GN=desV PE=1 SV=1 |
|
| 1.38e-70 | 391 | 756 | 3 | 363 | Erythromycin biosynthesis sensory transduction protein EryC1 OS=Saccharopolyspora erythraea (strain ATCC 11635 / DSM 40517 / JCM 4748 / NBRC 13426 / NCIMB 8594 / NRRL 2338) OX=405948 GN=eryC1 PE=3 SV=2 |
|
| 2.28e-70 | 390 | 755 | 2 | 364 | Pleiotropic regulatory protein OS=Geobacillus stearothermophilus OX=1422 GN=degT PE=3 SV=1 |
|
| 4.59e-58 | 409 | 759 | 21 | 371 | dTDP-3-amino-3,4,6-trideoxy-alpha-D-glucose transaminase OS=Micromonospora megalomicea subsp. nigra OX=136926 GN=megDII PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
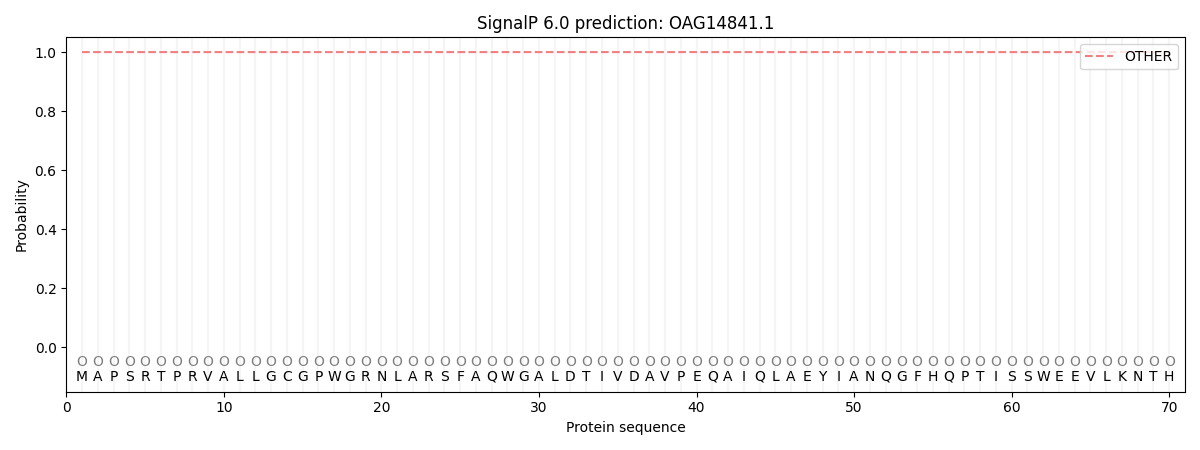
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000042 | 0.000000 |
