You are browsing environment: FUNGIDB
CAZyme Information: NEUTE1DRAFT_42678-t26_1-p1
You are here: Home > Sequence: NEUTE1DRAFT_42678-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora tetrasperma | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora tetrasperma | |||||||||||
| CAZyme ID | NEUTE1DRAFT_42678-t26_1-p1 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH131 | 22 | 260 | 1.6e-110 | 0.9882352941176471 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 408085 | GH131_N | 3.05e-74 | 21 | 250 | 1 | 246 | Glycoside hydrolase 131 catalytic N-terminal domain. This is the N-terminal domain found in glycoside hydrolase family 131 (GH131A) protein observed in Coprinopsis cinerea. GH131A exhibits bifunctional exo-beta-1,3-/-1,6- and endo-beta-1,4 activity toward beta-glucan. This domain is catalytic in nature though the catalytic mechanism of C. cinerea GH131A is different from that of typical glycosidases that use a pair of carboxylic acid residues as the catalytic residues. In the case of GH131A, Glu98 and His218 may form a catalytic dyad and Glu98 may activate His218 during catalysis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.16e-196 | 1 | 261 | 1 | 261 | |
| 1.81e-159 | 8 | 261 | 5 | 259 | |
| 5.52e-149 | 1 | 261 | 1 | 263 | |
| 1.80e-146 | 1 | 261 | 1 | 268 | |
| 1.80e-146 | 1 | 261 | 1 | 268 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.51e-136 | 20 | 261 | 2 | 243 | Chain A, Beta-glucanase [Podospora anserina],4LE3_B Chain B, Beta-glucanase [Podospora anserina],4LE3_C Chain C, Beta-glucanase [Podospora anserina],4LE3_D Chain D, Beta-glucanase [Podospora anserina],4LE4_A Chain A, Beta-glucanase [Podospora anserina],4LE4_B Chain B, Beta-glucanase [Podospora anserina],4LE4_C Chain C, Beta-glucanase [Podospora anserina],4LE4_D Chain D, Beta-glucanase [Podospora anserina] |
|
| 4.24e-77 | 21 | 259 | 3 | 232 | Chain A, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130],3W9A_B Chain B, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130],3W9A_C Chain C, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130],3W9A_D Chain D, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
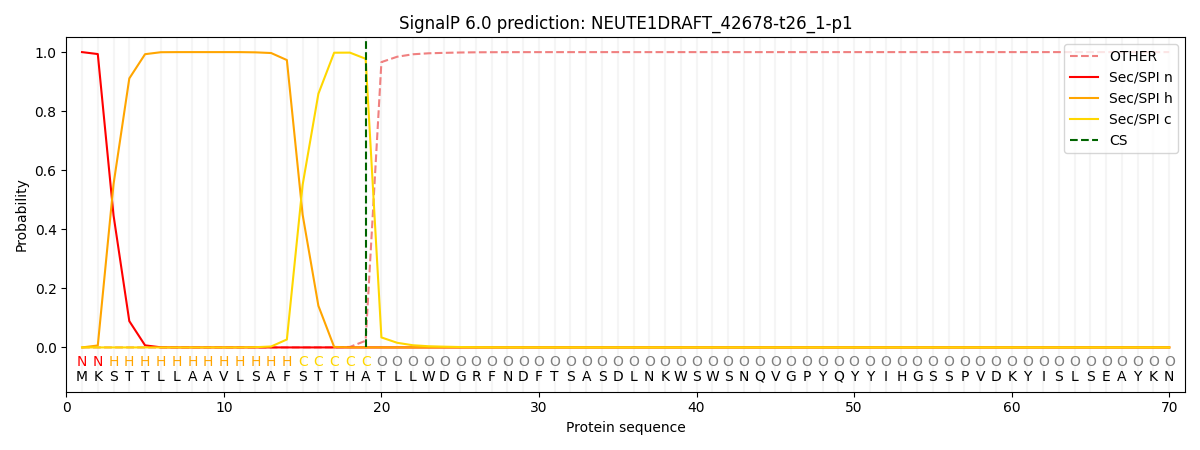
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000276 | 0.999701 | CS pos: 19-20. Pr: 0.9774 |
