You are browsing environment: FUNGIDB
CAZyme Information: NEUTE1DRAFT_37265-t26_1-p1
You are here: Home > Sequence: NEUTE1DRAFT_37265-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora tetrasperma | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora tetrasperma | |||||||||||
| CAZyme ID | NEUTE1DRAFT_37265-t26_1-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 616 | 837 | 2.7e-49 | 0.9847715736040609 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404513 | Glyco_trans_2_3 | 4.33e-48 | 616 | 826 | 1 | 194 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| 133045 | CESA_like | 5.86e-09 | 464 | 727 | 1 | 180 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| 224136 | BcsA | 2.35e-07 | 614 | 887 | 138 | 395 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| 177464 | PHA02682 | 7.22e-05 | 7 | 110 | 81 | 195 | ORF080 virion core protein; Provisional |
| 397323 | Atrophin-1 | 1.55e-04 | 11 | 111 | 172 | 270 | Atrophin-1 family. Atrophin-1 is the protein product of the dentatorubral-pallidoluysian atrophy (DRPLA) gene. DRPLA OMIM:125370 is a progressive neurodegenerative disorder. It is caused by the expansion of a CAG repeat in the DRPLA gene on chromosome 12p. This results in an extended polyglutamine region in atrophin-1, that is thought to confer toxicity to the protein, possibly through altering its interactions with other proteins. The expansion of a CAG repeat is also the underlying defect in six other neurodegenerative disorders, including Huntington's disease. One interaction of expanded polyglutamine repeats that is thought to be pathogenic is that with the short glutamine repeat in the transcriptional coactivator CREB binding protein, CBP. This interaction draws CBP away from its usual nuclear location to the expanded polyglutamine repeat protein aggregates that are characteristic of the polyglutamine neurodegenerative disorders. This interferes with CBP-mediated transcription and causes cytotoxicity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1001 | 1 | 1003 | |
| 0.0 | 248 | 1001 | 1 | 752 | |
| 0.0 | 109 | 1001 | 15 | 905 | |
| 0.0 | 107 | 1001 | 8 | 902 | |
| 0.0 | 91 | 1001 | 4 | 911 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000071 | 0.000000 |
TMHMM Annotations download full data without filtering help
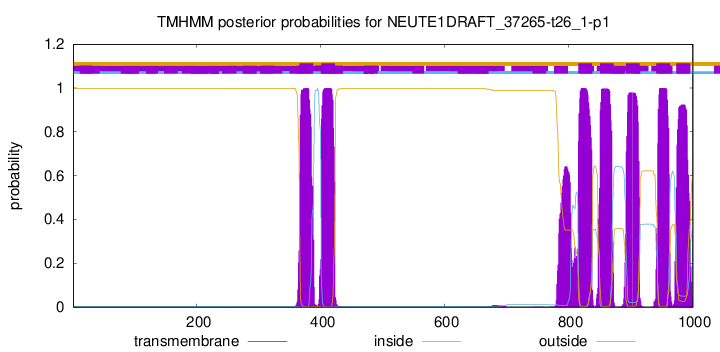
| Start | End |
|---|---|
| 366 | 388 |
| 401 | 423 |
| 816 | 838 |
| 851 | 873 |
| 893 | 915 |
| 942 | 964 |
| 974 | 996 |
