You are browsing environment: FUNGIDB
CAZyme Information: NEUTE1DRAFT_150113-t26_1-p1
Basic Information
help
| Species |
Neurospora tetrasperma
|
| Lineage |
Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora tetrasperma
|
| CAZyme ID |
NEUTE1DRAFT_150113-t26_1-p1
|
| CAZy Family |
GH26 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 603 |
|
66606.89 |
6.4132 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_NtetraspermaFGSC2508 |
10380 |
510951 |
0 |
10380
|
|
| Gene Location |
No EC number prediction in NEUTE1DRAFT_150113-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH76 |
69 |
499 |
9.2e-51 |
0.7513966480446927 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 397638
|
Glyco_hydro_76 |
4.97e-11 |
203 |
446 |
116 |
301 |
Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 5.07e-178 |
48 |
599 |
50 |
602 |
| 7.17e-178 |
2 |
599 |
3 |
602 |
| 7.17e-178 |
2 |
599 |
3 |
602 |
| 1.04e-177 |
1 |
599 |
1 |
624 |
| 7.60e-165 |
44 |
601 |
71 |
574 |
NEUTE1DRAFT_150113-t26_1-p1 has no PDB hit.
NEUTE1DRAFT_150113-t26_1-p1 has no Swissprot hit.
This protein is predicted as SP
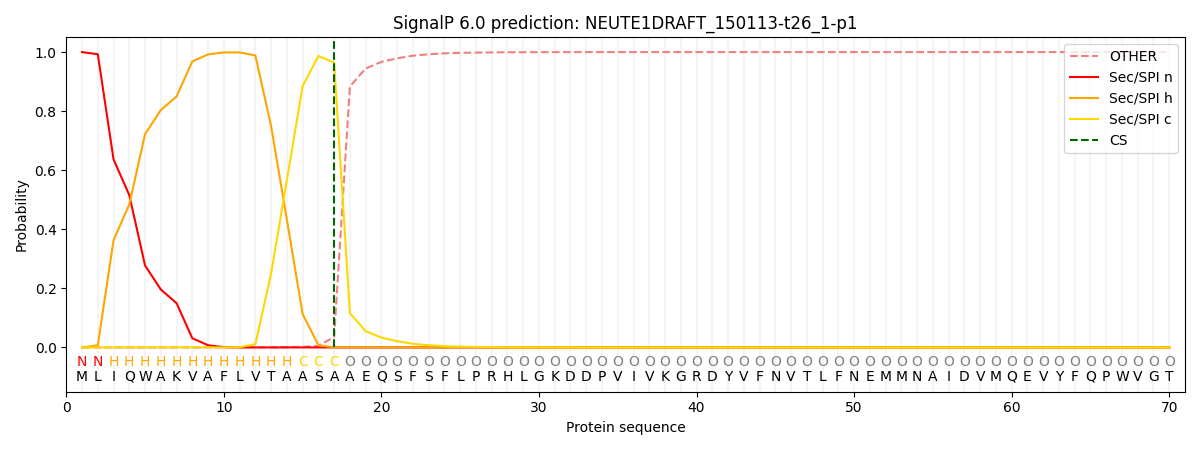
| Other |
SP_Sec_SPI |
CS Position |
| 0.000410 |
0.999579 |
CS pos: 17-18. Pr: 0.9643 |
There is no transmembrane helices in NEUTE1DRAFT_150113-t26_1-p1.

