You are browsing environment: FUNGIDB
CAZyme Information: NEUTE1DRAFT_147849-t26_1-p1
You are here: Home > Sequence: NEUTE1DRAFT_147849-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora tetrasperma | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora tetrasperma | |||||||||||
| CAZyme ID | NEUTE1DRAFT_147849-t26_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.164:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 29 | 481 | 2.6e-144 | 0.9956140350877193 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 405300 | Glyco_hydr_30_2 | 6.32e-16 | 30 | 353 | 6 | 357 | O-Glycosyl hydrolase family 30. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 488 | 1 | 488 | |
| 1.35e-249 | 22 | 488 | 19 | 487 | |
| 1.49e-246 | 1 | 488 | 1 | 487 | |
| 1.49e-246 | 1 | 488 | 1 | 487 | |
| 4.45e-241 | 19 | 488 | 20 | 487 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.41e-08 | 131 | 427 | 62 | 341 | Crystal Structure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.32e-206 | 22 | 488 | 18 | 478 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
|
| 7.20e-07 | 15 | 353 | 22 | 445 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
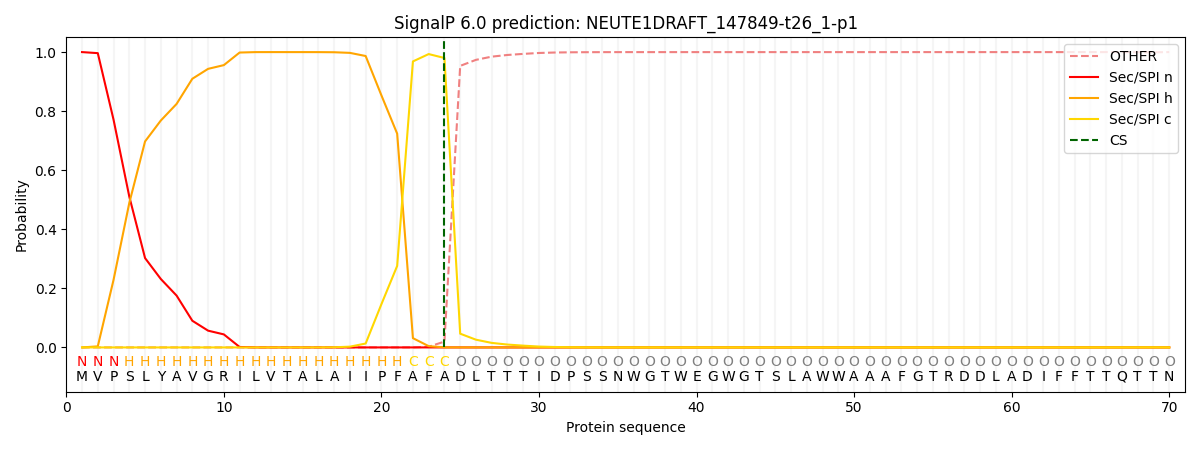
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000843 | 0.999146 | CS pos: 24-25. Pr: 0.9801 |
