You are browsing environment: FUNGIDB
CAZyme Information: NEUTE1DRAFT_132459-t26_1-p1
You are here: Home > Sequence: NEUTE1DRAFT_132459-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora tetrasperma | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora tetrasperma | |||||||||||
| CAZyme ID | NEUTE1DRAFT_132459-t26_1-p1 | |||||||||||
| CAZy Family | GH132 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 24 | 252 | 5.1e-18 | 0.6554054054054054 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119356 | GH18_hevamine_XipI_class_III | 1.28e-84 | 23 | 303 | 1 | 278 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
| 395573 | Glyco_hydro_18 | 4.90e-12 | 24 | 266 | 1 | 239 | Glycosyl hydrolases family 18. |
| 237865 | PRK14951 | 2.49e-09 | 1130 | 1255 | 369 | 491 | DNA polymerase III subunits gamma and tau; Provisional |
| 223021 | PHA03247 | 7.47e-09 | 1135 | 1378 | 2647 | 2881 | large tegument protein UL36; Provisional |
| 223021 | PHA03247 | 1.25e-08 | 1058 | 1381 | 2673 | 3004 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.70e-55 | 11 | 306 | 8 | 335 | |
| 1.25e-53 | 21 | 310 | 19 | 341 | |
| 9.24e-50 | 25 | 313 | 17 | 321 | |
| 9.28e-49 | 10 | 290 | 7 | 325 | |
| 9.28e-49 | 10 | 290 | 7 | 325 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.58e-37 | 27 | 287 | 5 | 286 | A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_B A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_C A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus] |
|
| 1.62e-37 | 27 | 287 | 6 | 287 | AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163],4TX6_B AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163] |
|
| 1.62e-37 | 27 | 287 | 6 | 287 | ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XTK_B ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XUC_A Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_B Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_C Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XVP_A ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163],2XVP_B ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163] |
|
| 2.20e-32 | 25 | 310 | 8 | 290 | ScCTS1_apo crystal structure [Saccharomyces cerevisiae],2UY3_A ScCTS1_8-chlorotheophylline crystal structure [Saccharomyces cerevisiae],2UY4_A ScCTS1_acetazolamide crystal structure [Saccharomyces cerevisiae],2UY5_A ScCTS1_kinetin crystal structure [Saccharomyces cerevisiae],4TXE_A ScCTS1 in complex with compound 5 [Saccharomyces cerevisiae] |
|
| 3.78e-24 | 27 | 288 | 5 | 256 | cDNA cloning and 1.75A crystal structure determination of PPL2, a novel chimerolectin from Parkia platycephala seeds exhibiting endochitinolytic activity [Parkia platycephala] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.07e-36 | 19 | 336 | 24 | 352 | Endochitinase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=ctcA PE=2 SV=1 |
|
| 3.68e-36 | 27 | 307 | 33 | 337 | Endochitinase 2 OS=Coccidioides posadasii (strain C735) OX=222929 GN=CTS2 PE=3 SV=2 |
|
| 4.26e-36 | 27 | 307 | 33 | 337 | Endochitinase 2 OS=Coccidioides immitis (strain RS) OX=246410 GN=CTS2 PE=3 SV=2 |
|
| 1.16e-34 | 4 | 336 | 7 | 360 | Endochitinase A1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=chiA1 PE=3 SV=1 |
|
| 3.73e-34 | 4 | 287 | 7 | 314 | Endochitinase A1 OS=Neosartorya fumigata OX=746128 GN=chiA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
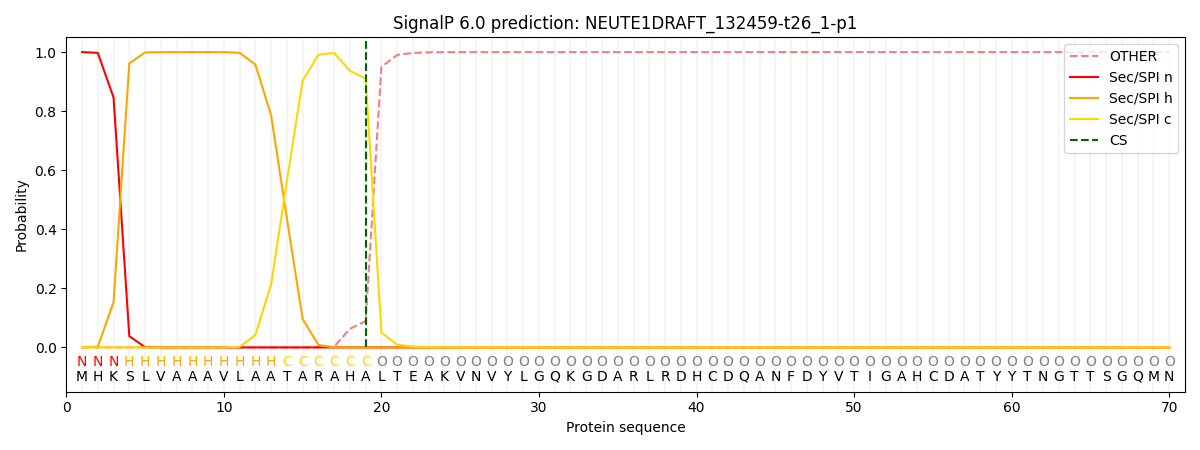
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000247 | 0.999724 | CS pos: 19-20. Pr: 0.9109 |
