You are browsing environment: FUNGIDB
CAZyme Information: NEUTE1DRAFT_119026-t26_1-p1
You are here: Home > Sequence: NEUTE1DRAFT_119026-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora tetrasperma | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora tetrasperma | |||||||||||
| CAZyme ID | NEUTE1DRAFT_119026-t26_1-p1 | |||||||||||
| CAZy Family | AA8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH132 | 132 | 439 | 1.5e-100 | 0.9834983498349835 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397780 | SUN | 3.24e-130 | 174 | 427 | 1 | 244 | Beta-glucosidase (SUN family). Members of this family include Nca3, Sun4 and Sim1. This is a family of yeast proteins, involved in a diverse set of functions (DNA replication, aging, mitochondrial biogenesis and cell septation). BGLA from Candida wickerhamii has been characterized as a Beta-glucosidase EC:3.2.1.21. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.86e-180 | 1 | 441 | 1 | 419 | |
| 7.81e-174 | 1 | 440 | 1 | 419 | |
| 3.00e-166 | 37 | 444 | 43 | 437 | |
| 6.03e-166 | 37 | 444 | 43 | 437 | |
| 6.03e-166 | 37 | 444 | 43 | 437 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.28e-78 | 5 | 433 | 6 | 407 | Secreted beta-glucosidase sun1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=sun1 PE=1 SV=1 |
|
| 3.40e-78 | 2 | 433 | 5 | 440 | Probable secreted beta-glucosidase ARB_04747 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_04747 PE=1 SV=1 |
|
| 5.21e-68 | 171 | 433 | 162 | 412 | Probable secreted beta-glucosidase SUN4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SUN4 PE=1 SV=1 |
|
| 6.40e-66 | 167 | 433 | 214 | 468 | Probable secreted beta-glucosidase SIM1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SIM1 PE=1 SV=2 |
|
| 1.07e-64 | 171 | 433 | 105 | 355 | Probable secreted beta-glucosidase UTH1 OS=Saccharomyces cerevisiae (strain RM11-1a) OX=285006 GN=UTH1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
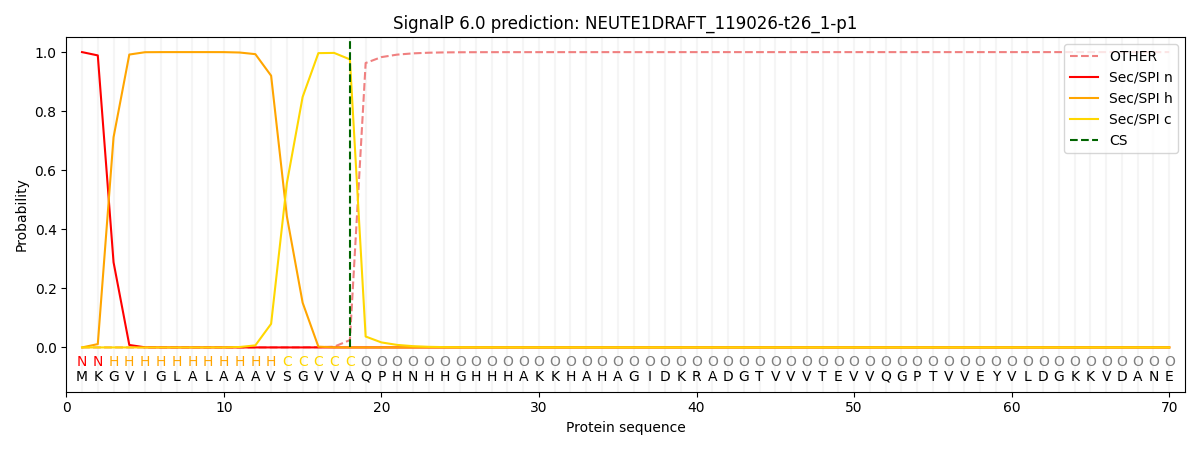
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000260 | 0.999713 | CS pos: 18-19. Pr: 0.9753 |
