You are browsing environment: FUNGIDB
CAZyme Information: NEUDI_82221T0-p1
You are here: Home > Sequence: NEUDI_82221T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora discreta | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora discreta | |||||||||||
| CAZyme ID | NEUDI_82221T0-p1 | |||||||||||
| CAZy Family | GT15 | |||||||||||
| CAZyme Description | 1,3-beta-glucan synthase/callose synthase catalytic subunit | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:56 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 859 | 1641 | 0 | 0.9878213802435724 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 859 | 1691 | 1 | 819 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 2.19e-48 | 344 | 453 | 1 | 111 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 340863 | MFS_MefA_like | 0.002 | 483 | 644 | 194 | 357 | Macrolide efflux protein A and similar proteins of the Major Facilitator Superfamily of transporters. This family is composed of Streptococcus pyogenes macrolide efflux protein A (MefA) and similar transporters, many of which remain uncharacterized. Some members may be multidrug resistance (MDR) transporters, which are drug/H+ antiporters (DHAs) that mediate the efflux of a variety of drugs and toxic compounds, conferring resistance to these compounds. MefA confers resistance to 14-membered macrolides including erythromycin and to 15-membered macrolides. It functions as an efflux pump to regulate intracellular macrolide levels. The MefA-like family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 111 | 1938 | 134 | 1943 | |
| 0.0 | 111 | 1938 | 134 | 1943 | |
| 0.0 | 62 | 1956 | 62 | 1932 | |
| 0.0 | 111 | 1938 | 132 | 1941 | |
| 0.0 | 97 | 1925 | 99 | 1907 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 198 | 1885 | 15 | 1739 | 1,3-beta-glucan synthase component FKS3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS3 PE=1 SV=1 |
|
| 0.0 | 107 | 1906 | 82 | 1888 | 1,3-beta-glucan synthase component FKS1 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=fksA PE=3 SV=1 |
|
| 0.0 | 113 | 1886 | 96 | 1847 | 1,3-beta-glucan synthase component GSC2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GSC2 PE=1 SV=2 |
|
| 0.0 | 139 | 1878 | 84 | 1781 | 1,3-beta-glucan synthase component FKS1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=FKS1 PE=3 SV=3 |
|
| 0.0 | 115 | 1891 | 85 | 1836 | 1,3-beta-glucan synthase component FKS1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000034 | 0.000014 |
TMHMM Annotations download full data without filtering help
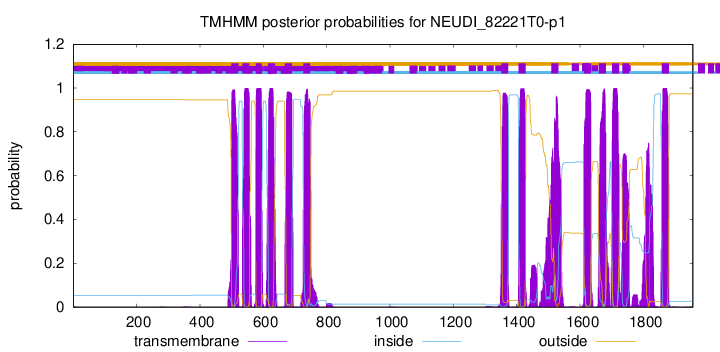
| Start | End |
|---|---|
| 499 | 521 |
| 534 | 556 |
| 576 | 595 |
| 615 | 637 |
| 668 | 690 |
| 726 | 748 |
| 1351 | 1373 |
| 1406 | 1428 |
| 1509 | 1531 |
| 1612 | 1634 |
| 1659 | 1681 |
| 1701 | 1723 |
| 1733 | 1755 |
| 1858 | 1880 |
