You are browsing environment: FUNGIDB
CAZyme Information: NEUDI_79750T0-p1
You are here: Home > Sequence: NEUDI_79750T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora discreta | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora discreta | |||||||||||
| CAZyme ID | NEUDI_79750T0-p1 | |||||||||||
| CAZy Family | GH92 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA12 | 26 | 434 | 7.7e-176 | 0.9950124688279302 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225044 | YliI | 1.12e-18 | 31 | 402 | 46 | 380 | Glucose/arabinose dehydrogenase, beta-propeller fold [Carbohydrate transport and metabolism]. |
| 293791 | 7WD40 | 0.002 | 55 | 174 | 177 | 289 | WD40 repeats in seven bladed beta propellers. The WD40 repeat is found in a number of eukaryotic proteins that cover a wide variety of functions including adaptor/regulatory modules in signal transduction, pre-mRNA processing, and cytoskeleton assembly. It typically contains a GH dipeptide 11-24 residues from its N-terminus and the WD dipeptide at its C-terminus and is 40 residues long, hence the name WD40. Between the GH and WD dipeptides lies a conserved core. It forms a propeller-like structure with several blades where each blade is composed of a four-stranded anti-parallel beta-sheet. The WD40 sequence repeat originally described in literature forms the first three strands of one blade and the last strand in the next blade. The C-terminal WD40 repeat completes the blade structure of the N-terminal WD40 repeat to create the closed ring propeller-structure. The residues on the top and bottom surface of the propeller are proposed to coordinate interactions with other proteins and/or small ligands, allowing them to bind either stably or reversibly. |
| 225921 | YvrE | 0.003 | 104 | 141 | 167 | 206 | Sugar lactone lactonase YvrE [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.41e-299 | 1 | 507 | 1 | 515 | |
| 9.51e-183 | 16 | 501 | 14 | 480 | |
| 2.34e-171 | 16 | 501 | 14 | 482 | |
| 1.39e-157 | 12 | 446 | 10 | 439 | |
| 3.73e-155 | 16 | 501 | 14 | 483 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.67e-132 | 32 | 436 | 7 | 402 | Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 1.72e-132 | 32 | 436 | 8 | 403 | Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 1.78e-132 | 32 | 436 | 9 | 404 | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 1.60e-67 | 24 | 436 | 4 | 410 | Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_A Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_B Chain B, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
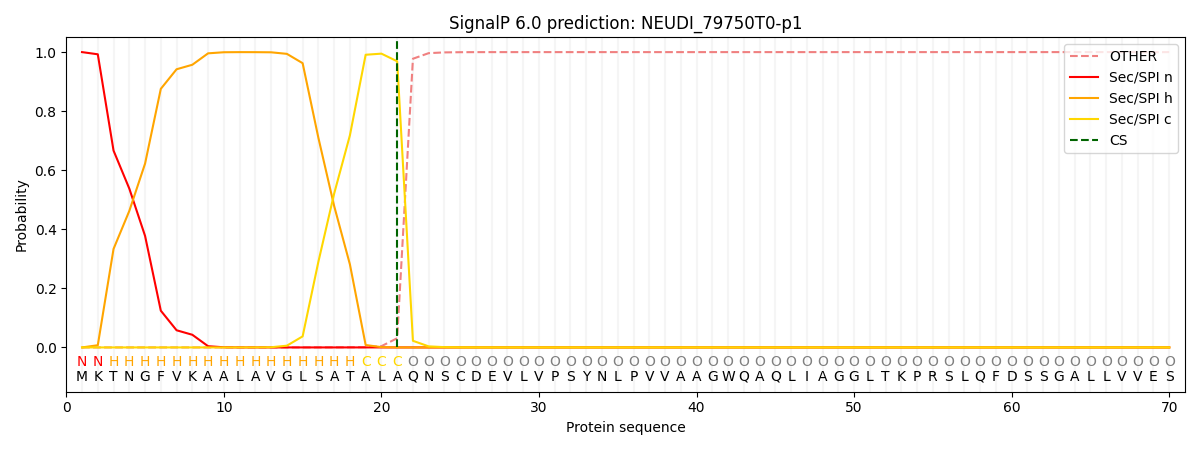
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000462 | 0.999519 | CS pos: 21-22. Pr: 0.9689 |
