You are browsing environment: FUNGIDB
CAZyme Information: NEUDI_59781T0-p1
Basic Information
help
| Species |
Neurospora discreta
|
| Lineage |
Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora discreta
|
| CAZyme ID |
NEUDI_59781T0-p1
|
| CAZy Family |
GH6 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_NdiscretaFGSC8579 |
10345 |
510953 |
397 |
9948
|
|
| Gene Location |
No EC number prediction in NEUDI_59781T0-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH12 |
140 |
277 |
2.6e-26 |
0.8589743589743589 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 396303
|
Glyco_hydro_12 |
4.99e-11 |
26 |
278 |
9 |
206 |
Glycosyl hydrolase family 12. |
| 235746
|
PRK06215 |
1.82e-10 |
50 |
274 |
50 |
233 |
hypothetical protein; Provisional |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 3.76e-100 |
29 |
286 |
17 |
275 |
| 3.76e-100 |
29 |
286 |
17 |
275 |
| 7.56e-100 |
29 |
286 |
17 |
275 |
| 4.67e-88 |
35 |
315 |
1 |
277 |
| 7.17e-67 |
24 |
270 |
11 |
234 |
NEUDI_59781T0-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.71e-17 |
37 |
278 |
40 |
256 |
Endoglucanase cel12C OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel12C PE=3 SV=1 |
| 4.33e-14 |
33 |
271 |
25 |
233 |
Xyloglucan-specific endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgeA PE=1 SV=1 |
This protein is predicted as SP
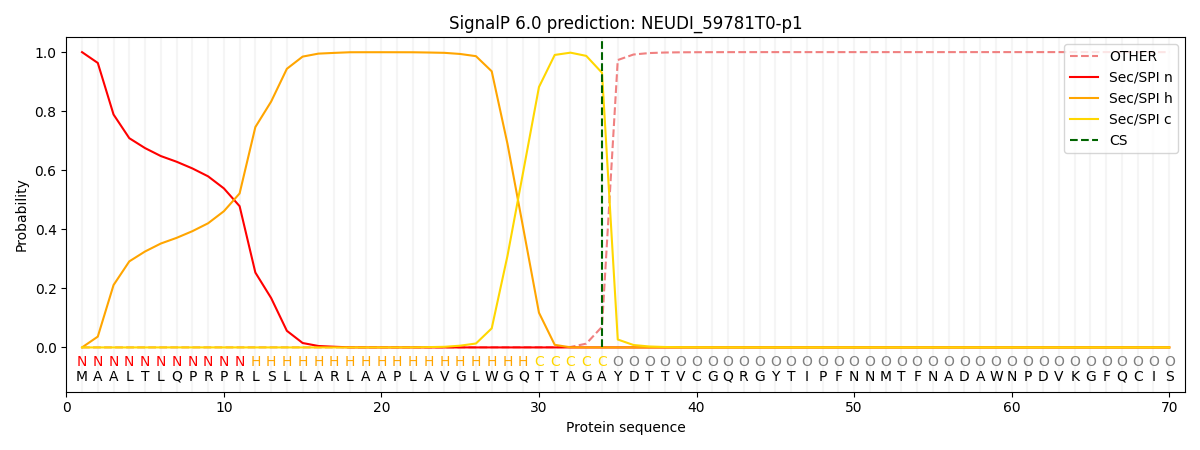
| Other |
SP_Sec_SPI |
CS Position |
| 0.000735 |
0.999231 |
CS pos: 34-35. Pr: 0.9300 |
There is no transmembrane helices in NEUDI_59781T0-p1.

