You are browsing environment: FUNGIDB
CAZyme Information: NEUDI_20779T0-p1
Basic Information
help
| Species |
Neurospora discreta
|
| Lineage |
Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora discreta
|
| CAZyme ID |
NEUDI_20779T0-p1
|
| CAZy Family |
GH43 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 275 |
|
31272.30 |
6.6397 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_NdiscretaFGSC8579 |
10345 |
510953 |
397 |
9948
|
|
| Gene Location |
No EC number prediction in NEUDI_20779T0-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH26 |
85 |
225 |
3.7e-22 |
0.44224422442244227 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 396639
|
Glyco_hydro_26 |
9.96e-09 |
94 |
193 |
139 |
244 |
Glycosyl hydrolase family 26. |
| 226609
|
ManB2 |
3.16e-04 |
104 |
242 |
178 |
336 |
Beta-mannanase [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 2.19e-52 |
11 |
260 |
39 |
298 |
| 4.47e-52 |
11 |
264 |
43 |
300 |
| 8.67e-52 |
11 |
270 |
39 |
307 |
| 4.39e-51 |
11 |
264 |
90 |
347 |
| 1.35e-50 |
11 |
270 |
39 |
307 |
NEUDI_20779T0-p1 has no PDB hit.
NEUDI_20779T0-p1 has no Swissprot hit.
This protein is predicted as OTHER
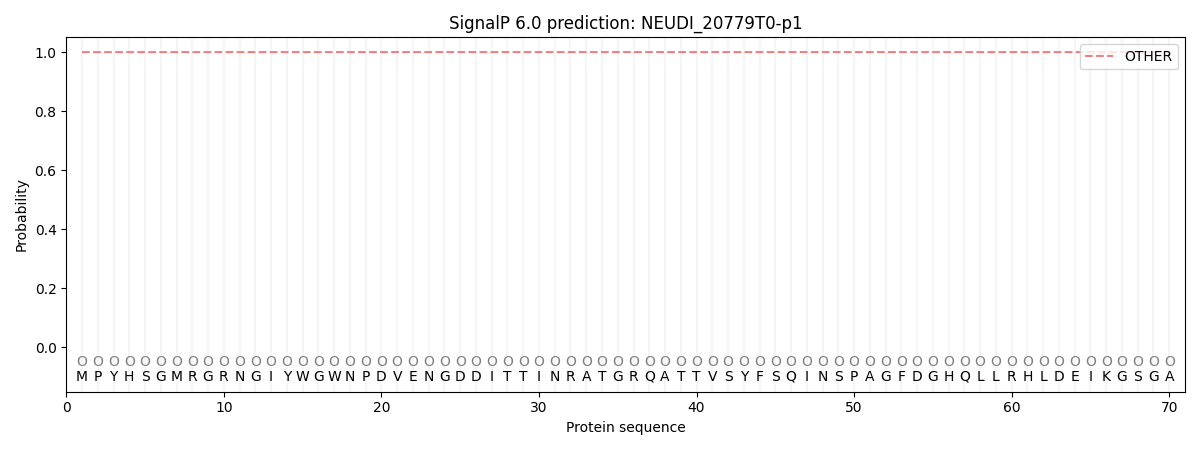
| Other |
SP_Sec_SPI |
CS Position |
| 1.000044 |
0.000000 |
|
There is no transmembrane helices in NEUDI_20779T0-p1.

