You are browsing environment: FUNGIDB
NEUDI_166551T0-p1
Basic Information
help
Species
Neurospora discreta
Lineage
Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora discreta
CAZyme ID
NEUDI_166551T0-p1
CAZy Family
GH3
CAZyme Description
unspecified product
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
468
53339.27
7.7087
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_NdiscretaFGSC8579
10345
510953
397
9948
Gene Location
No EC number prediction in NEUDI_166551T0-p1.
Family
Start
End
Evalue
family coverage
GT34
114
369
4.4e-27
0.926829268292683
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
368535
Glyco_transf_34
5.68e-12
134
277
38
190
galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34.
more
215616
PLN03181
8.96e-04
176
250
192
265
glycosyltransferase; Provisional
more
215617
PLN03182
0.006
162
318
180
327
xyloglucan 6-xylosyltransferase; Provisional
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.76e-46
105
404
145
386
1.76e-46
105
404
145
386
1.76e-46
105
404
145
386
1.76e-46
105
404
145
386
1.65e-44
105
407
208
454
NEUDI_166551T0-p1 has no PDB hit.
NEUDI_166551T0-p1 has no Swissprot hit.
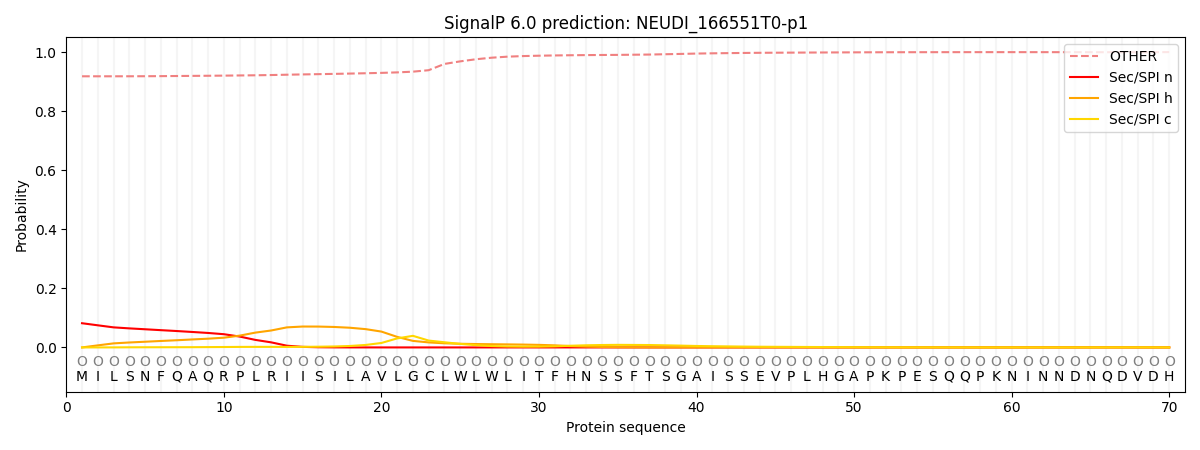
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
0.918629
0.081372