You are browsing environment: FUNGIDB
CAZyme Information: NEUDI_163849T0-p1
You are here: Home > Sequence: NEUDI_163849T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora discreta | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora discreta | |||||||||||
| CAZyme ID | NEUDI_163849T0-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 745 | 946 | 4.6e-41 | 0.39956331877729256 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238220 | RNase_T2 | 1.09e-15 | 88 | 212 | 2 | 123 | Ribonuclease T2 (RNase T2) is a widespread family of secreted RNases found in every organism examined thus far. This family includes RNase Rh, RNase MC1, RNase LE, and self-incompatibility RNases (S-RNases). Plant T2 RNases are expressed during leaf senescence in order to scavenge phosphate from ribonucleotides. They are also expressed in response to wounding or pathogen invasion. S-RNases are thought to prevent self-fertilization by acting as selective cytotoxins of "self" pollen. |
| 238512 | RNase_T2_euk | 1.42e-14 | 88 | 212 | 12 | 124 | Ribonuclease T2 (RNase T2) is a widespread family of secreted RNases found in every organism examined thus far. This family includes RNase Rh, RNase MC1, RNase LE, and self-incompatibility RNases (S-RNases). Plant T2 RNases are expressed during leaf senescence in order to scavenge phosphate from ribonucleotides. They are also expressed in response to wounding or pathogen invasion. S-RNases are thought to prevent self-fertilization by acting as selective cytotoxins of "self" pollen. Generally, RNases have two distinct binding sites: the primary site (B1 site) and the subsite (B2 site), for nucleotides located at the 5'- and 3'- terminal ends of the sessil bond, respectively. This CD includes the eukaryotic RNase T2 family members. |
| 396238 | FAD_binding_4 | 2.76e-14 | 755 | 890 | 1 | 129 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 395357 | Ribonuclease_T2 | 3.58e-12 | 101 | 196 | 26 | 109 | Ribonuclease T2 family. |
| 223354 | GlcD | 4.63e-10 | 755 | 889 | 32 | 159 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.13e-17 | 755 | 941 | 598 | 775 | |
| 4.13e-17 | 755 | 941 | 598 | 775 | |
| 3.27e-12 | 724 | 941 | 39 | 238 | |
| 3.71e-11 | 755 | 955 | 48 | 232 | |
| 2.39e-10 | 755 | 955 | 20 | 204 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.82e-20 | 755 | 938 | 124 | 297 | Crystal structure of VAO-type flavoprotein MtVAO615 at pH 7.5 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_A Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_B Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464] |
|
| 5.64e-14 | 733 | 940 | 23 | 211 | The crystal structure of EncM H138T mutant [Streptomyces maritimus],6FYE_B The crystal structure of EncM H138T mutant [Streptomyces maritimus] |
|
| 1.31e-13 | 733 | 940 | 23 | 211 | The crystal structure of EncM T139V mutant [Streptomyces maritimus],6FYD_B The crystal structure of EncM T139V mutant [Streptomyces maritimus],6FYD_C The crystal structure of EncM T139V mutant [Streptomyces maritimus],6FYD_D The crystal structure of EncM T139V mutant [Streptomyces maritimus] |
|
| 2.30e-13 | 733 | 940 | 23 | 211 | The crystal structure of EncM V135M mutant [Streptomyces maritimus],6FYF_B The crystal structure of EncM V135M mutant [Streptomyces maritimus],6FYF_C The crystal structure of EncM V135M mutant [Streptomyces maritimus],6FYF_D The crystal structure of EncM V135M mutant [Streptomyces maritimus] |
|
| 3.04e-13 | 733 | 940 | 23 | 211 | The crystal structure of EncM V135T mutant [Streptomyces maritimus],6FYG_B The crystal structure of EncM V135T mutant [Streptomyces maritimus],6FYG_C The crystal structure of EncM V135T mutant [Streptomyces maritimus],6FYG_D The crystal structure of EncM V135T mutant [Streptomyces maritimus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.20e-23 | 767 | 1229 | 144 | 562 | FAD-linked oxidoreductase OXR2 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=OXR2 PE=2 SV=1 |
|
| 1.42e-23 | 767 | 950 | 135 | 309 | FAD-linked oxidoreductase ffsJ OS=Aspergillus flavipes OX=41900 GN=ffsJ PE=3 SV=1 |
|
| 4.25e-22 | 645 | 1229 | 29 | 548 | FAD-linked oxidoreductase ZEB1 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=ZEB1 PE=2 SV=2 |
|
| 6.45e-22 | 645 | 939 | 27 | 302 | FAD-linked oxidoreductase malF OS=Malbranchea aurantiaca OX=78605 GN=malF PE=1 SV=1 |
|
| 2.12e-21 | 766 | 1246 | 139 | 591 | FAD-linked oxidoreductase easE OS=Epichloe festucae var. lolii OX=73839 GN=easE PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
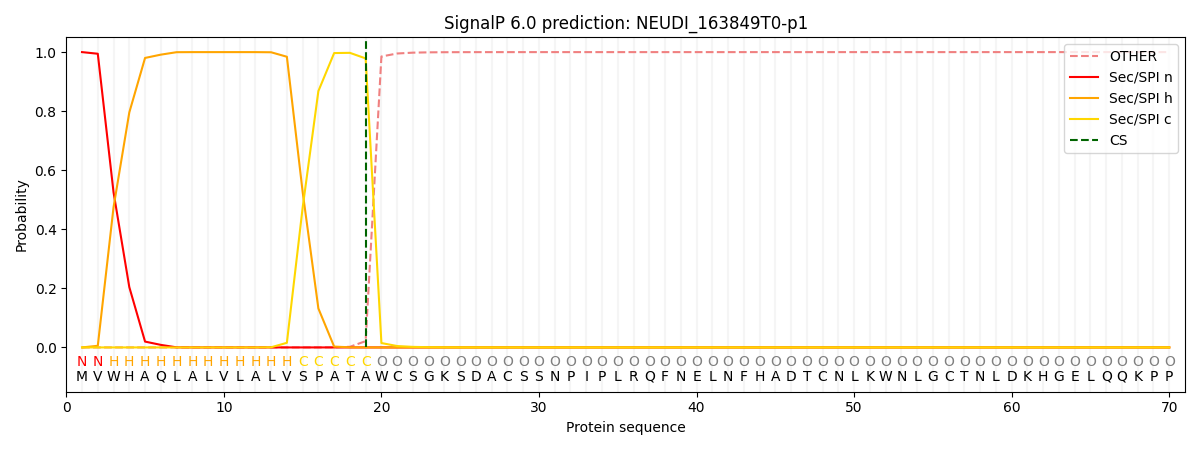
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000201 | 0.999767 | CS pos: 19-20. Pr: 0.9786 |
