You are browsing environment: FUNGIDB
CAZyme Information: NEUDI_136208T0-p1
You are here: Home > Sequence: NEUDI_136208T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora discreta | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora discreta | |||||||||||
| CAZyme ID | NEUDI_136208T0-p1 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | RNA-binding protein NOVA1/PASILLA and related KH domain proteins | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:61 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 46 | 221 | 5.8e-76 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 6.74e-106 | 46 | 220 | 1 | 175 | Glycosyl hydrolases family 11. |
| 197593 | fCBD | 4.57e-12 | 262 | 295 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 5.64e-11 | 263 | 291 | 1 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.71e-185 | 1 | 295 | 1 | 293 | |
| 1.62e-147 | 1 | 295 | 1 | 287 | |
| 1.62e-147 | 1 | 295 | 1 | 287 | |
| 1.62e-147 | 1 | 295 | 1 | 287 | |
| 9.96e-144 | 1 | 292 | 1 | 356 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.21e-113 | 36 | 226 | 2 | 193 | Chain A, endoxylanase 11A [Thermochaetoides thermophila],1XNK_B Chain B, endoxylanase 11A [Thermochaetoides thermophila] |
|
| 1.44e-112 | 36 | 224 | 2 | 191 | Chain A, Endo-1,4-beta-xylanase [Thermochaetoides thermophila],1H1A_B Chain B, Endo-1,4-beta-xylanase [Thermochaetoides thermophila] |
|
| 2.59e-98 | 42 | 223 | 6 | 188 | Chain A, Endo-1,4-beta-xylanase 2 [Trichoderma reesei],4XQD_B Chain B, Endo-1,4-beta-xylanase 2 [Trichoderma reesei],5ZF3_A Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose [Trichoderma reesei RUT C-30],5ZH0_A Crystal Structures of Endo-beta-1,4-xylanase II [Trichoderma reesei RUT C-30],5ZO0_A Neutron structure of xylanase at pD5.4 [Trichoderma reesei RUT C-30] |
|
| 2.68e-98 | 42 | 223 | 7 | 189 | Chain A, Endo-1,4-beta-xylanase 2 [Trichoderma reesei] |
|
| 2.68e-98 | 42 | 223 | 7 | 189 | High resolution structure of xylanase-II from one micron beam experiment [Trichoderma longibrachiatum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.26e-103 | 1 | 223 | 1 | 222 | Endo-1,4-beta-xylanase 2 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=xyn2 PE=1 SV=1 |
|
| 1.26e-103 | 1 | 223 | 1 | 222 | Endo-1,4-beta-xylanase 2 OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=xyn2 PE=1 SV=2 |
|
| 1.23e-102 | 1 | 295 | 1 | 290 | Endo-1,4-beta-xylanase B OS=Phanerodontia chrysosporium OX=2822231 GN=xynB PE=1 SV=1 |
|
| 1.58e-101 | 1 | 223 | 1 | 220 | Endo-1,4-beta-xylanase 2 OS=Trichoderma harzianum OX=5544 GN=Xyn2 PE=1 SV=1 |
|
| 3.64e-99 | 1 | 295 | 1 | 310 | Endo-1,4-beta-xylanase B OS=Penicillium oxalicum OX=69781 GN=xynB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
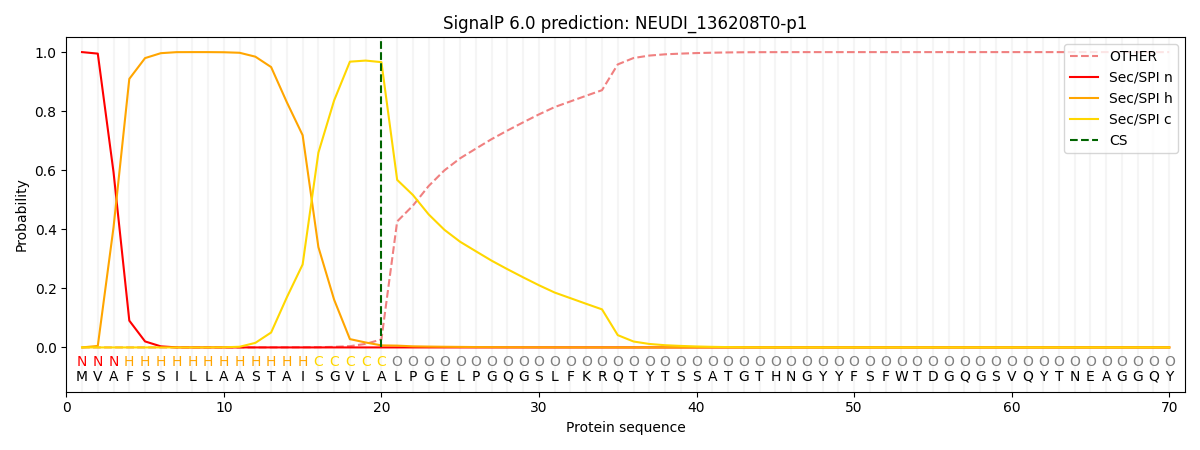
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000415 | 0.999555 | CS pos: 20-21. Pr: 0.9660 |
