You are browsing environment: FUNGIDB
CAZyme Information: NEUDI_103987T0-p1
Basic Information
help
| Species |
Neurospora discreta
|
| Lineage |
Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora discreta
|
| CAZyme ID |
NEUDI_103987T0-p1
|
| CAZy Family |
AA2 |
| CAZyme Description |
Nucleolar GTPase/ATPase p130
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1071 |
|
118099.05 |
6.6971 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_NdiscretaFGSC8579 |
10345 |
510953 |
397 |
9948
|
|
| Gene Location |
No EC number prediction in NEUDI_103987T0-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GT90 |
549 |
769 |
3.5e-48 |
0.756 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 214773
|
CAP10 |
0.001 |
569 |
767 |
29 |
195 |
Putative lipopolysaccharide-modifying enzyme. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
1070 |
1 |
1092 |
| 1.86e-209 |
29 |
770 |
49 |
765 |
| 5.41e-205 |
11 |
770 |
11 |
760 |
| 1.37e-202 |
11 |
770 |
31 |
760 |
| 7.23e-201 |
31 |
770 |
53 |
767 |
NEUDI_103987T0-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.12e-27 |
333 |
776 |
152 |
599 |
Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
This protein is predicted as OTHER
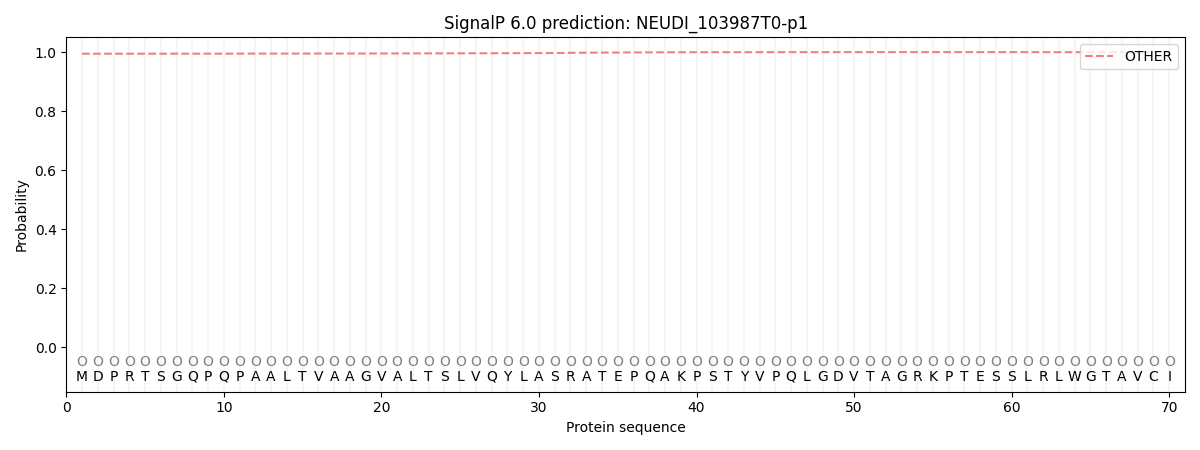
| Other |
SP_Sec_SPI |
CS Position |
| 0.994963 |
0.005053 |
|
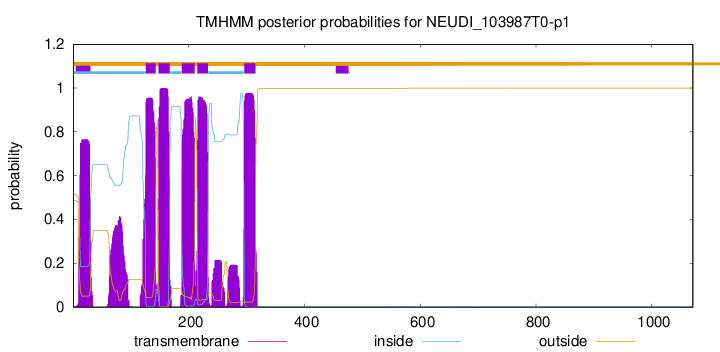
| Start |
End |
| 126 |
143 |
| 148 |
167 |
| 188 |
210 |
| 215 |
233 |
| 296 |
315 |


