You are browsing environment: FUNGIDB
CAZyme Information: NCU08473-t26_1-p1
You are here: Home > Sequence: NCU08473-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora crassa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora crassa | |||||||||||
| CAZyme ID | NCU08473-t26_1-p1 | |||||||||||
| CAZy Family | GH93 | |||||||||||
| CAZyme Description | alpha-1,3-glucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.59:2 | - | - |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH71 | 21 | 409 | 7.7e-132 | 0.968 |
| CBM24 | 454 | 534 | 6.7e-26 | 0.9868421052631579 |
| CBM24 | 553 | 630 | 1.7e-24 | 0.9868421052631579 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397634 | Glyco_hydro_71 | 1.59e-144 | 21 | 409 | 1 | 363 | Glycosyl hydrolase family 71. Family of alpha-1,3-glucanases. |
| 211418 | GH71 | 6.20e-113 | 15 | 307 | 1 | 283 | Glycoside hydrolase family 71. This family of glycoside hydrolases 71 (following the CAZY nomenclature) function as alpha-1,3-glucanases (mutanases, EC 3.2.1.59). They appear to have an endo-hydrolytic mode of enzymatic activity and bacterial members are investigated as candidates for the development of dental caries treatments.The member from fission yeast, endo-alpha-1,3-glucanase Agn1p, plays a vital role in daughter cell separation, while Agn2p has been associated with endolysis of the ascus wall. |
| 211414 | GH99_GH71_like | 7.29e-33 | 21 | 298 | 1 | 280 | Glycoside hydrolase families 71, 99, and related domains. This superfamily of glycoside hydrolases contains families GH71 and GH99 (following the CAZY nomenclature), as well as other members with undefined function and specificity. |
| 173787 | Peptidases_S8_S53 | 6.15e-12 | 1182 | 1427 | 1 | 236 | Peptidase domain in the S8 and S53 families. Members of the peptidases S8 (subtilisin and kexin) and S53 (sedolisin) family include endopeptidases and exopeptidases. The S8 family has an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. Serine acts as a nucleophile, aspartate as an electrophile, and histidine as a base. The S53 family contains a catalytic triad Glu/Asp/Ser with an additional acidic residue Asp in the oxyanion hole, similar to that of subtilisin. The serine residue here is the nucleophilic equivalent of the serine residue in the S8 family, while glutamic acid has the same role here as the histidine base. However, the aspartic acid residue that acts as an electrophile is quite different. In S53, it follows glutamic acid, while in S8 it precedes histidine. The stability of these enzymes may be enhanced by calcium; some members have been shown to bind up to 4 ions via binding sites with different affinity. There is a great diversity in the characteristics of their members: some contain disulfide bonds, some are intracellular while others are extracellular, some function at extreme temperatures, and others at high or low pH values. |
| 395035 | Peptidase_S8 | 1.02e-08 | 1179 | 1429 | 1 | 268 | Subtilase family. Subtilases are a family of serine proteases. They appear to have independently and convergently evolved an Asp/Ser/His catalytic triad, like that found in the trypsin serine proteases (see pfam00089). Structure is an alpha/beta fold containing a 7-stranded parallel beta sheet, order 2314567. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1736 | 1 | 1736 | |
| 0.0 | 1 | 889 | 1 | 880 | |
| 4.31e-185 | 9 | 563 | 9 | 568 | |
| 1.60e-176 | 9 | 527 | 9 | 533 | |
| 2.35e-176 | 3 | 715 | 4 | 730 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.28e-55 | 14 | 441 | 31 | 432 | Mutanase Pc12g07500 OS=Penicillium rubens (strain ATCC 28089 / DSM 1075 / NRRL 1951 / Wisconsin 54-1255) OX=500485 GN=PCH_Pc12g07500 PE=1 SV=1 |
|
| 1.56e-42 | 19 | 401 | 22 | 384 | Glucan endo-1,3-alpha-glucosidase agn1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=agn1 PE=1 SV=2 |
|
| 8.63e-23 | 19 | 330 | 11 | 328 | Ascus wall endo-1,3-alpha-glucanase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=agn2 PE=1 SV=2 |
|
| 3.45e-09 | 1144 | 1417 | 118 | 365 | Alkaline protease 1 OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=alp1 PE=3 SV=1 |
|
| 1.32e-07 | 1144 | 1438 | 117 | 380 | Alkaline protease 1 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=alp1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
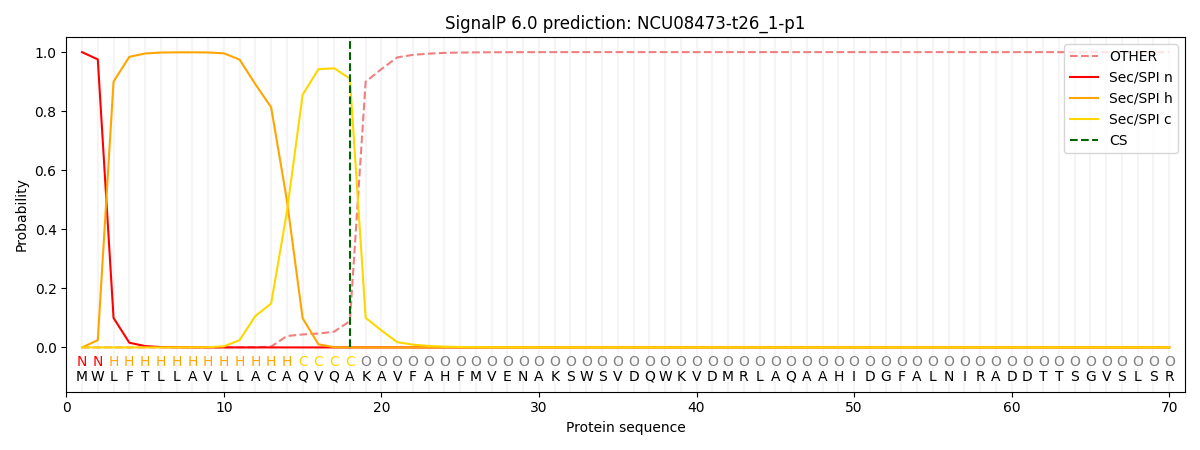
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000815 | 0.999178 | CS pos: 18-19. Pr: 0.9104 |
