You are browsing environment: FUNGIDB
CAZyme Information: NCU08087-t26_1-p1
Basic Information
help
| Species |
Neurospora crassa
|
| Lineage |
Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora crassa
|
| CAZyme ID |
NCU08087-t26_1-p1
|
| CAZy Family |
GH76 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 311 |
CM002236|CGC21 |
35397.26 |
6.7421 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_NcrassaOR74A |
10591 |
367110 |
833 |
9758
|
|
| Gene Location |
No EC number prediction in NCU08087-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH26 |
119 |
261 |
1.2e-22 |
0.44884488448844884 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 396639
|
Glyco_hydro_26 |
1.10e-09 |
130 |
229 |
139 |
244 |
Glycosyl hydrolase family 26. |
| 226609
|
ManB2 |
5.36e-05 |
140 |
278 |
178 |
336 |
Beta-mannanase [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 9.72e-52 |
47 |
300 |
43 |
300 |
| 2.64e-51 |
47 |
308 |
39 |
309 |
| 9.59e-51 |
47 |
300 |
90 |
347 |
| 1.46e-50 |
47 |
308 |
39 |
309 |
| 2.26e-49 |
47 |
308 |
39 |
309 |
NCU08087-t26_1-p1 has no PDB hit.
NCU08087-t26_1-p1 has no Swissprot hit.
This protein is predicted as SP
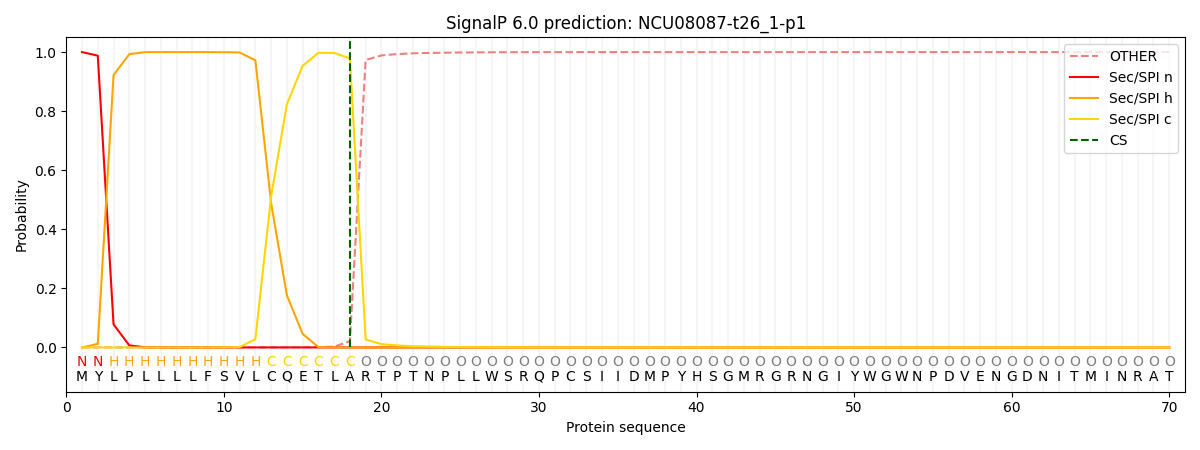
| Other |
SP_Sec_SPI |
CS Position |
| 0.000235 |
0.999735 |
CS pos: 18-19. Pr: 0.9790 |
There is no transmembrane helices in NCU08087-t26_1-p1.

