You are browsing environment: FUNGIDB
CAZyme Information: NCU07817-t26_1-p1
Basic Information
help
| Species |
Neurospora crassa
|
| Lineage |
Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora crassa
|
| CAZyme ID |
NCU07817-t26_1-p1
|
| CAZy Family |
GH71|CBM24|CBM24 |
| CAZyme Description |
non-anchored cell wall protein 3
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 266 |
|
27515.06 |
5.0352 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_NcrassaOR74A |
10591 |
367110 |
833 |
9758
|
|
| Gene Location |
No EC number prediction in NCU07817-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| CBM52 |
41 |
90 |
4.3e-29 |
0.9423076923076923 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 402331
|
Carb_bind |
4.21e-20 |
42 |
89 |
2 |
49 |
Carbohydrate binding. This is a carbohydrate binding domain which has been shown in Schizosaccharomyces pombe to be required for septum localization. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 9.22e-201 |
1 |
266 |
1 |
266 |
| 2.33e-123 |
18 |
266 |
1 |
247 |
| 5.46e-122 |
18 |
266 |
1 |
247 |
| 5.18e-120 |
18 |
266 |
1 |
247 |
| 1.11e-112 |
18 |
266 |
1 |
249 |
NCU07817-t26_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 7.44e-17 |
111 |
266 |
41 |
195 |
Uncharacterized secreted protein ARB_07637 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07637 PE=1 SV=1 |
This protein is predicted as SP
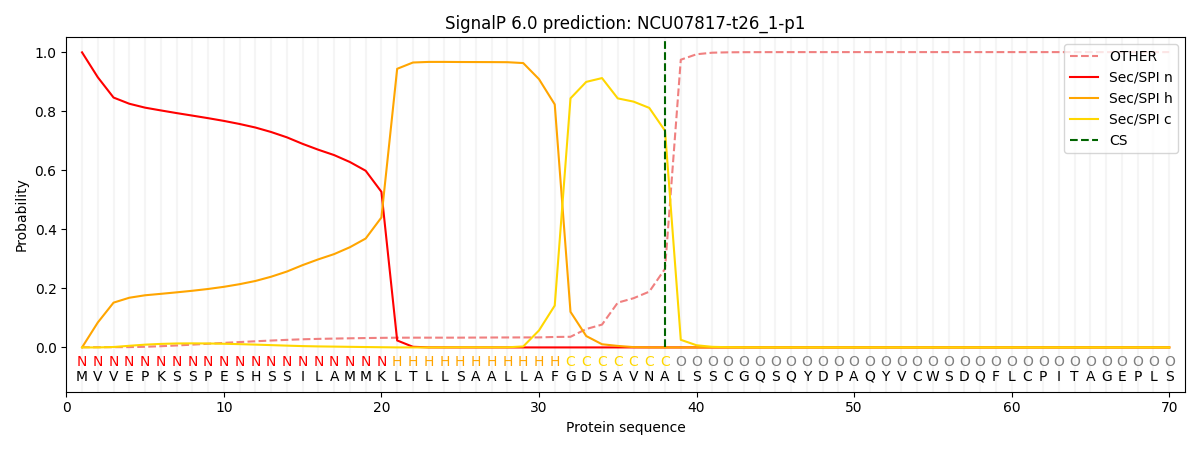
| Other |
SP_Sec_SPI |
CS Position |
| 0.001250 |
0.998735 |
CS pos: 38-39. Pr: 0.7336 |
There is no transmembrane helices in NCU07817-t26_1-p1.

