You are browsing environment: FUNGIDB
CAZyme Information: NCU05105-t26_1-p1
You are here: Home > Sequence: NCU05105-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora crassa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora crassa | |||||||||||
| CAZyme ID | NCU05105-t26_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | glucan endo-1,3-beta-glucosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.39:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 41 | 780 | 1.4e-214 | 0.9851351351351352 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403800 | Pectate_lyase_3 | 1.05e-35 | 82 | 289 | 17 | 213 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 239169 | MopB_NADH-Q-OR-NuoG2 | 7.94e-04 | 373 | 461 | 122 | 204 | MopB_NADH-Q-OR-NuoG2: The NuoG/Nad11/75-kDa subunit (second domain) of the NADH-quinone oxidoreductase (NADH-Q-OR)/respiratory complex I/NADH dehydrogenase-1 (NDH-1). The NADH-Q-OR is the first energy-transducting complex in the respiratory chains of many prokaryotes and eukaryotes. Mitochondrial complex I and its bacterial counterpart, NDH-1, function as a redox pump that uses the redox energy to translocate H+ ions across the membrane, resulting in a significant contribution to energy production. The atomic structure of complex I is not known and the mechanisms of electron transfer and proton pumping are not established. The nad11 gene codes for the largest (75-kDa) subunit of the mitochondrial NADH:ubiquinone oxidoreductase, it constitutes the electron input part of the enzyme, or the so-called NADH dehydrogenase fragment. In Escherichia coli, this subunit is encoded by the nuoG gene, and is part of the 14 distinct subunits constituting the 'minimal' functional enzyme. The nad11 gene is nuclear-encoded in animals, plants, and fungi, but is still encoded in the mitochondrial genome of some protists. The Nad11/NuoG subunit is made of two domains: the first contains three binding sites for FeS clusters (the fer2 domain), the second domain (this CD), is of unknown function or, as postulated, has lost an ancestral formate dehydrogenase activity that became redundant during the evolution of the complex I enzyme. Although only vestigial sequence evidence remains of a molybdopterin binding site, this protein domain family belongs to the molybdopterin_binding (MopB) superfamily of proteins. Bacterial type II NADH-quinone oxidoreductases and NQR-type sodium-motive NADH-quinone oxidoreductases are not homologs of this domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 787 | 1 | 787 | |
| 0.0 | 1 | 787 | 1 | 798 | |
| 0.0 | 45 | 784 | 37 | 767 | |
| 0.0 | 39 | 784 | 18 | 766 | |
| 0.0 | 40 | 784 | 31 | 761 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.02e-73 | 44 | 748 | 3 | 723 | Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila],5M60_A Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila] |
|
| 1.29e-65 | 47 | 750 | 26 | 727 | Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQN_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_A Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.16e-297 | 6 | 783 | 3 | 761 | Glucan endo-1,3-beta-glucosidase BGN13.1 OS=Trichoderma harzianum OX=5544 GN=bgn13.1 PE=1 SV=1 |
|
| 3.33e-63 | 60 | 744 | 56 | 750 | Glucan 1,3-beta-glucosidase OS=Cochliobolus carbonum OX=5017 GN=EXG1 PE=1 SV=1 |
|
| 1.23e-59 | 38 | 748 | 34 | 843 | Probable glucan endo-1,3-beta-glucosidase ARB_02077 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02077 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
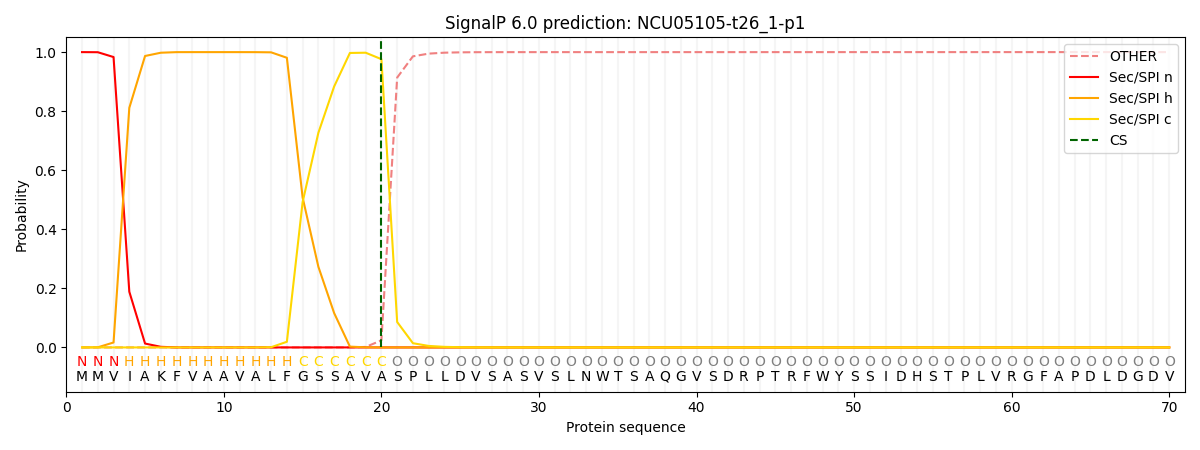
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000238 | 0.999707 | CS pos: 20-21. Pr: 0.9761 |
