You are browsing environment: FUNGIDB
CAZyme Information: NCU03646-t26_1-p1
You are here: Home > Sequence: NCU03646-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora crassa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora crassa | |||||||||||
| CAZyme ID | NCU03646-t26_1-p1 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA2 | 64 | 243 | 2.4e-25 | 0.6823529411764706 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 173829 | plant_peroxidase_like_1 | 2.54e-159 | 24 | 289 | 1 | 264 | Uncharacterized family of plant peroxidase-like proteins. This is a subgroup of heme-dependent peroxidases similar to plant peroxidases. Along with animal peroxidases, these enzymes belong to a group of peroxidases containing a heme prosthetic group (ferriprotoporphyrin IX) which catalyzes a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. The plant peroxidase-like superfamily is found in all three kingdoms of life and carries out a variety of biosynthetic and degradative functions. |
| 396406 | WSC | 3.44e-26 | 621 | 699 | 3 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 173823 | plant_peroxidase_like | 8.49e-24 | 65 | 288 | 18 | 254 | Heme-dependent peroxidases similar to plant peroxidases. Along with animal peroxidases, these enzymes belong to a group of peroxidases containing a heme prosthetic group (ferriprotoporphyrin IX), which catalyzes a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. The plant peroxidase-like superfamily is found in all three kingdoms of life and carries out a variety of biosynthetic and degradative functions. Several sub-families can be identified. Class I includes intracellular peroxidases present in fungi, plants, archaea and bacteria, called catalase-peroxidases, that can exhibit both catalase and broad-spectrum peroxidase activities depending on the steady-state concentration of hydrogen peroxide. Catalase-peroxidases are typically comprised of two homologous domains that probably arose via a single gene duplication event. Class II includes ligninase and other extracellular fungal peroxidases, while class III is comprised of classic extracellular plant peroxidases, like horseradish peroxidase. |
| 395089 | peroxidase | 2.54e-21 | 64 | 200 | 15 | 157 | Peroxidase. |
| 396406 | WSC | 1.72e-20 | 743 | 821 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 8 | 836 | 11 | 803 | |
| 7.09e-281 | 14 | 833 | 16 | 804 | |
| 6.57e-175 | 22 | 833 | 23 | 736 | |
| 2.83e-103 | 6 | 532 | 3 | 535 | |
| 1.76e-101 | 6 | 498 | 3 | 500 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.20e-170 | 20 | 828 | 19 | 715 | WSC domain-containing protein ARB_07870 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07870 PE=1 SV=1 |
|
| 2.03e-35 | 611 | 829 | 12 | 220 | Putative fungistatic metabolite OS=Chaetomium globosum (strain ATCC 6205 / CBS 148.51 / DSM 1962 / NBRC 6347 / NRRL 1970) OX=306901 GN=CHGG_05463 PE=1 SV=2 |
|
| 3.04e-22 | 616 | 828 | 63 | 286 | WSC domain-containing protein ARB_07867 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07867 PE=1 SV=1 |
|
| 3.77e-12 | 621 | 829 | 140 | 329 | WSC domain-containing protein 2 OS=Danio rerio OX=7955 GN=wscd2 PE=3 SV=1 |
|
| 7.83e-11 | 621 | 829 | 133 | 322 | WSC domain-containing protein 2 OS=Homo sapiens OX=9606 GN=WSCD2 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
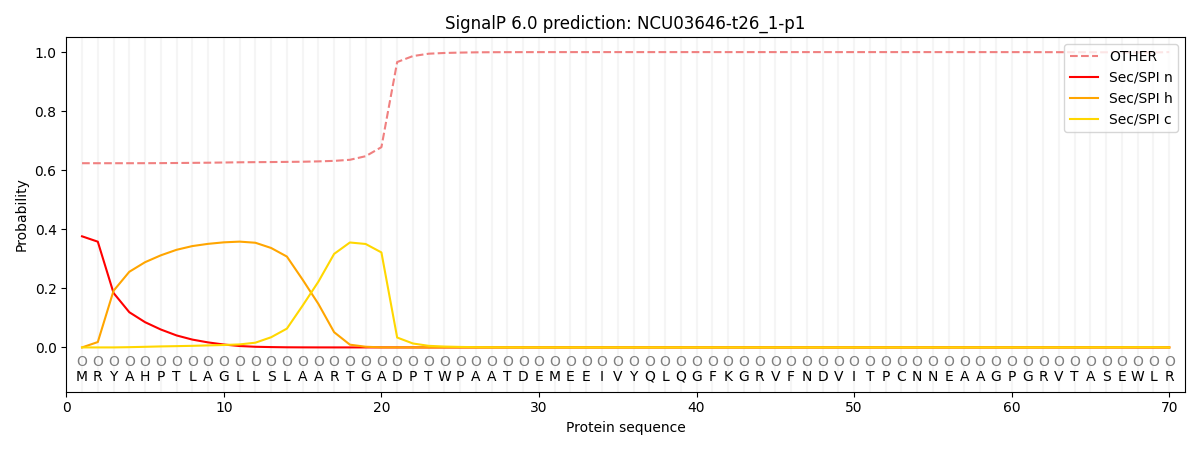
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.639146 | 0.360862 |
