You are browsing environment: FUNGIDB
CAZyme Information: NCU03215-t26_1-p1
Basic Information
help
| Species |
Neurospora crassa
|
| Lineage |
Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora crassa
|
| CAZyme ID |
NCU03215-t26_1-p1
|
| CAZy Family |
CE4 |
| CAZyme Description |
glycosyl hydrolase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 638 |
CM002236|CGC1 |
70463.10 |
6.1346 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_NcrassaOR74A |
10591 |
367110 |
833 |
9758
|
|
| Gene Location |
No EC number prediction in NCU03215-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH76 |
69 |
508 |
1.2e-51 |
0.7821229050279329 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 397638
|
Glyco_hydro_76 |
1.34e-09 |
203 |
446 |
116 |
301 |
Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 8.40e-194 |
45 |
634 |
47 |
602 |
| 1.24e-193 |
1 |
634 |
1 |
624 |
| 9.59e-193 |
2 |
634 |
3 |
602 |
| 9.59e-193 |
2 |
634 |
3 |
602 |
| 2.57e-179 |
45 |
634 |
66 |
611 |
NCU03215-t26_1-p1 has no PDB hit.
NCU03215-t26_1-p1 has no Swissprot hit.
This protein is predicted as SP
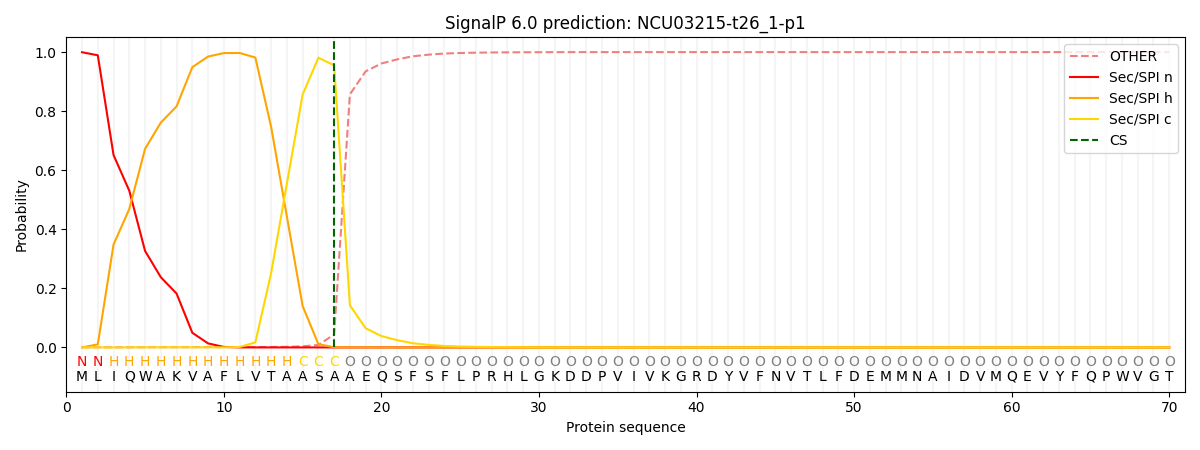
| Other |
SP_Sec_SPI |
CS Position |
| 0.001217 |
0.998778 |
CS pos: 17-18. Pr: 0.9555 |
There is no transmembrane helices in NCU03215-t26_1-p1.

