You are browsing environment: FUNGIDB
CAZyme Information: NCU01380-t26_1-p1
You are here: Home > Sequence: NCU01380-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neurospora crassa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Sordariaceae; Neurospora; Neurospora crassa | |||||||||||
| CAZyme ID | NCU01380-t26_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.53:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 20 | 214 | 3.5e-76 | 0.9685863874345549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397484 | Glyco_hydro_61 | 0.002 | 106 | 174 | 109 | 182 | Glycosyl hydrolase family 61. Although weak endoglucanase activity has been demonstrated in several members of this family, they lack the clustered conserved catalytic acidic amino acids present in most glycoside hydrolases. Many members of this family lack measurable cellulase activity on their own, but enhance the activity of other cellulolytic enzymes. They are therefore unlikely to be true glycoside hydrolases. The subsrate-binding surface of this family is a flat Ig-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.52e-234 | 1 | 413 | 1 | 413 | |
| 1.26e-84 | 11 | 412 | 9 | 420 | |
| 2.10e-84 | 3 | 412 | 2 | 401 | |
| 2.31e-84 | 3 | 403 | 2 | 412 | |
| 9.25e-84 | 9 | 411 | 47 | 465 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.14e-60 | 20 | 217 | 1 | 204 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
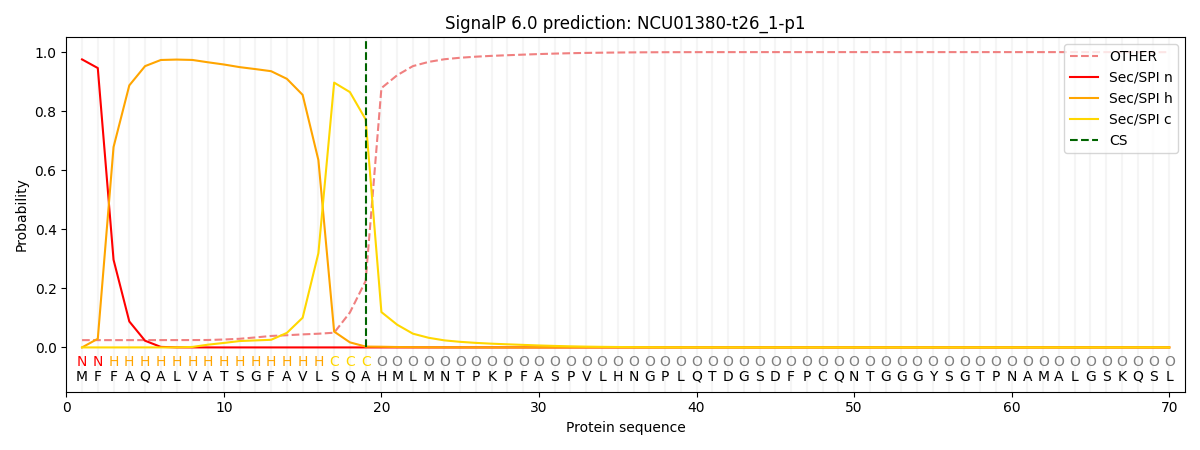
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.029743 | 0.970243 | CS pos: 19-20. Pr: 0.7721 |
