You are browsing environment: FUNGIDB
CAZyme Information: MELLADRAFT_92297-t26_1-p1
You are here: Home > Sequence: MELLADRAFT_92297-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
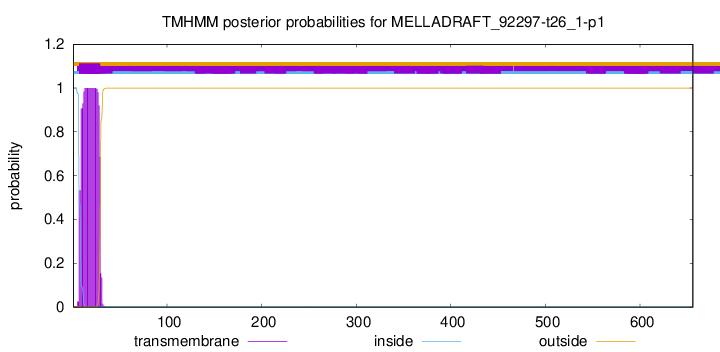
TMHMM annotations
Basic Information help
| Species | Melampsora larici-populina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Melampsoraceae; Melampsora; Melampsora larici-populina | |||||||||||
| CAZyme ID | MELLADRAFT_92297-t26_1-p1 | |||||||||||
| CAZy Family | GT66 | |||||||||||
| CAZyme Description | family 90 glycosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214773 | CAP10 | 1.42e-21 | 501 | 644 | 123 | 254 | Putative lipopolysaccharide-modifying enzyme. |
| 310354 | Glyco_transf_90 | 1.95e-12 | 524 | 643 | 215 | 323 | Glycosyl transferase family 90. This family of glycosyl transferases are specifically (mannosyl) glucuronoxylomannan/galactoxylomannan -beta 1,2-xylosyltransferases, EC:2.4.2.-. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.88e-190 | 107 | 645 | 148 | 692 | |
| 1.65e-170 | 106 | 656 | 194 | 716 | |
| 1.11e-120 | 116 | 654 | 235 | 752 | |
| 4.93e-115 | 124 | 647 | 114 | 636 | |
| 1.75e-110 | 124 | 643 | 133 | 640 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.45e-10 | 514 | 643 | 226 | 342 | human POGLUT1 in complex with Notch1 EGF12 and UDP [Homo sapiens],5L0S_A human POGLUT1 in complex with Factor VII EGF1 and UDP [Homo sapiens],5L0T_A human POGLUT1 in complex with EGF(+) and UDP [Homo sapiens],5L0U_A human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose [Homo sapiens],5L0V_A human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP [Homo sapiens],5UB5_A human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP [Homo sapiens] |
|
| 9.37e-08 | 514 | 645 | 261 | 374 | Crystal structure of Drosophila Poglut1 (Rumi) complexed with its glycoprotein product (glucosylated EGF repeat) and UDP [Drosophila melanogaster],5F85_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) and UDP [Drosophila melanogaster],5F86_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) [Drosophila melanogaster],5F87_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_B Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_C Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_D Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_E Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_F Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.06e-98 | 123 | 654 | 151 | 667 | Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
|
| 2.38e-10 | 480 | 643 | 241 | 389 | O-glucosyltransferase rumi homolog OS=Culex quinquefasciatus OX=7176 GN=CPIJ013394 PE=3 SV=1 |
|
| 3.00e-10 | 514 | 643 | 254 | 370 | Protein O-glucosyltransferase 1 OS=Rattus norvegicus OX=10116 GN=Poglut1 PE=3 SV=1 |
|
| 2.91e-09 | 514 | 643 | 254 | 370 | Protein O-glucosyltransferase 1 OS=Bos taurus OX=9913 GN=POGLUT1 PE=2 SV=1 |
|
| 3.98e-09 | 523 | 644 | 360 | 468 | Protein O-glucosyltransferase 2 OS=Homo sapiens OX=9606 GN=POGLUT2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
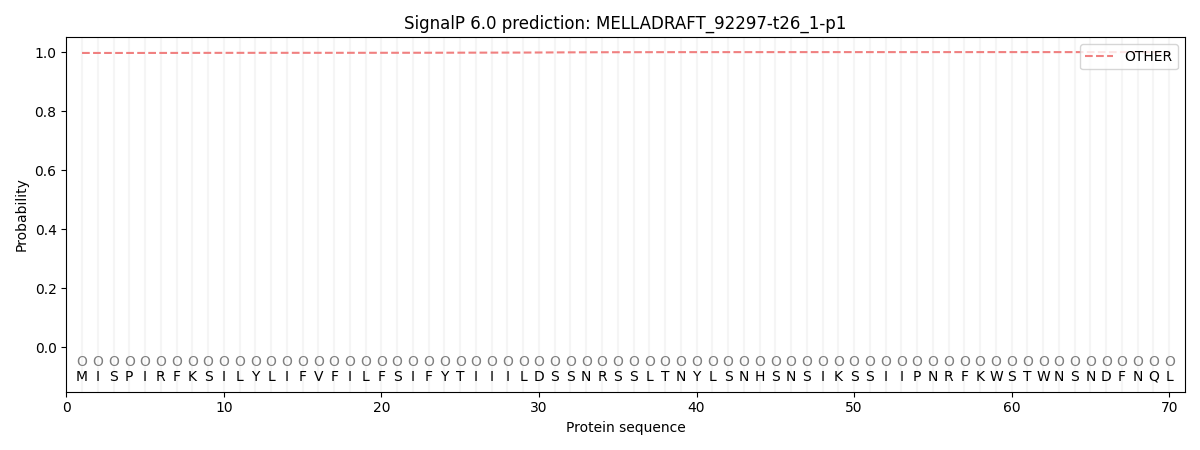
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.997253 | 0.002742 |