You are browsing environment: FUNGIDB
CAZyme Information: MELLADRAFT_78608-t26_1-p1
You are here: Home > Sequence: MELLADRAFT_78608-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Melampsora larici-populina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Melampsoraceae; Melampsora; Melampsora larici-populina | |||||||||||
| CAZyme ID | MELLADRAFT_78608-t26_1-p1 | |||||||||||
| CAZy Family | GH7 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA9 | 83 | 251 | 3.9e-27 | 0.7318181818181818 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 410622 | LPMO_AA9 | 7.88e-29 | 97 | 265 | 48 | 216 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 9 (AA9). AA9 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs) involved in the cleavage of cellulose chains with oxidation of carbons C1 and/or C4 and C6. Activities include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54) and lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56). The family used to be called GH61 because weak endoglucanase activity had been demonstrated in some family members. |
| 397484 | Glyco_hydro_61 | 9.15e-21 | 107 | 249 | 58 | 193 | Glycosyl hydrolase family 61. Although weak endoglucanase activity has been demonstrated in several members of this family, they lack the clustered conserved catalytic acidic amino acids present in most glycoside hydrolases. Many members of this family lack measurable cellulase activity on their own, but enhance the activity of other cellulolytic enzymes. They are therefore unlikely to be true glycoside hydrolases. The subsrate-binding surface of this family is a flat Ig-like fold. |
| 399033 | CFEM | 8.67e-08 | 348 | 414 | 5 | 66 | CFEM domain. This fungal specific cysteine rich domain is found in some proteins with proposed roles in fungal pathogenesis. The structure of the CFEM domain containing protein 'Surface antigen protein 2' from Candida albicans has been solved. |
| 128986 | CFEM | 3.76e-06 | 348 | 414 | 5 | 65 | eight cysteine-containing domain present in fungal extracellular membrane proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.17e-77 | 32 | 413 | 28 | 388 | |
| 9.34e-19 | 4 | 253 | 3 | 232 | |
| 4.56e-15 | 98 | 251 | 79 | 227 | |
| 5.77e-15 | 99 | 251 | 51 | 195 | |
| 2.14e-14 | 111 | 271 | 82 | 230 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.56e-10 | 126 | 266 | 106 | 248 | Chain A, Cel61b [Trichoderma reesei],2VTC_B Chain B, Cel61b [Trichoderma reesei] |
|
| 3.26e-07 | 129 | 264 | 78 | 219 | Chain A, LPMO9F [Malbranchea cinnamomea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.72e-11 | 105 | 253 | 90 | 228 | Cellulose-growth-specific protein OS=Agaricus bisporus OX=5341 GN=cel1 PE=3 SV=1 |
|
| 1.19e-10 | 28 | 277 | 31 | 255 | Polysaccharide monooxygenase Cel61a OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Cel61a PE=1 SV=1 |
|
| 4.40e-09 | 126 | 266 | 106 | 248 | Endoglucanase-7 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cel61b PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
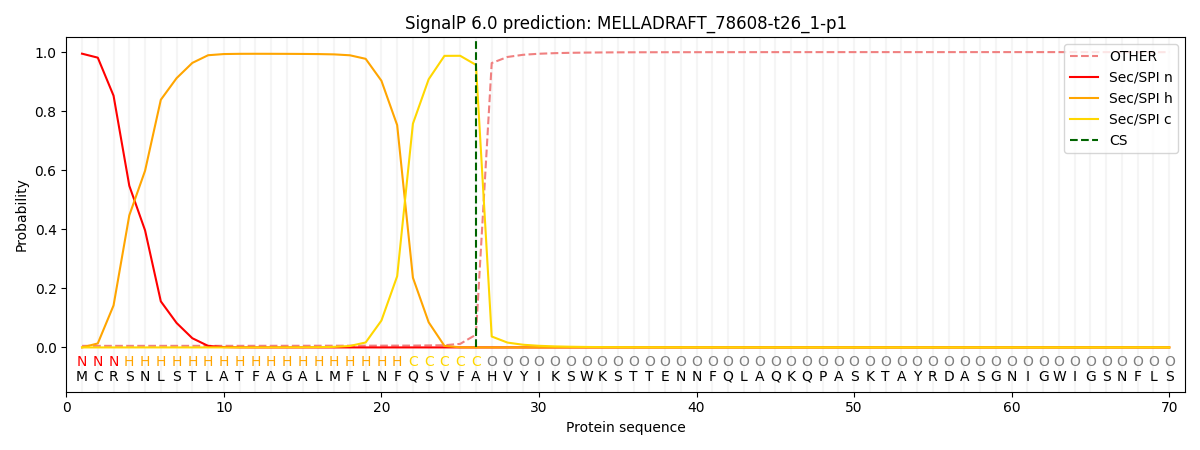
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.008157 | 0.991791 | CS pos: 26-27. Pr: 0.9567 |
