You are browsing environment: FUNGIDB
CAZyme Information: MELLADRAFT_78333-t26_1-p1
You are here: Home > Sequence: MELLADRAFT_78333-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Melampsora larici-populina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Melampsoraceae; Melampsora; Melampsora larici-populina | |||||||||||
| CAZyme ID | MELLADRAFT_78333-t26_1-p1 | |||||||||||
| CAZy Family | GH7 | |||||||||||
| CAZyme Description | family 5 glycoside hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.78:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 102 | 265 | 3.5e-23 | 0.56640625 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.97e-118 | 137 | 437 | 6 | 341 | |
| 1.26e-68 | 66 | 488 | 49 | 479 | |
| 1.34e-54 | 323 | 606 | 44 | 317 | |
| 2.07e-30 | 66 | 477 | 22 | 410 | |
| 6.62e-29 | 66 | 486 | 60 | 473 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.37e-07 | 152 | 371 | 96 | 249 | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
|
| 6.41e-07 | 152 | 371 | 82 | 235 | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.89e-07 | 66 | 257 | 54 | 227 | Mannan endo-1,4-beta-mannosidase 2 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN2 PE=2 SV=2 |
|
| 7.96e-07 | 65 | 257 | 38 | 211 | Mannan endo-1,4-beta-mannosidase 5 OS=Solanum lycopersicum OX=4081 GN=MAN5 PE=2 SV=1 |
|
| 7.96e-07 | 65 | 257 | 38 | 211 | Mannan endo-1,4-beta-mannosidase 2 OS=Solanum lycopersicum OX=4081 GN=MAN2 PE=2 SV=2 |
|
| 4.25e-06 | 63 | 322 | 26 | 233 | Mannan endo-1,4-beta-mannosidase 1 OS=Arabidopsis thaliana OX=3702 GN=MAN1 PE=2 SV=1 |
|
| 5.43e-06 | 65 | 257 | 26 | 199 | Mannan endo-1,4-beta-mannosidase 1 OS=Solanum lycopersicum OX=4081 GN=MAN1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
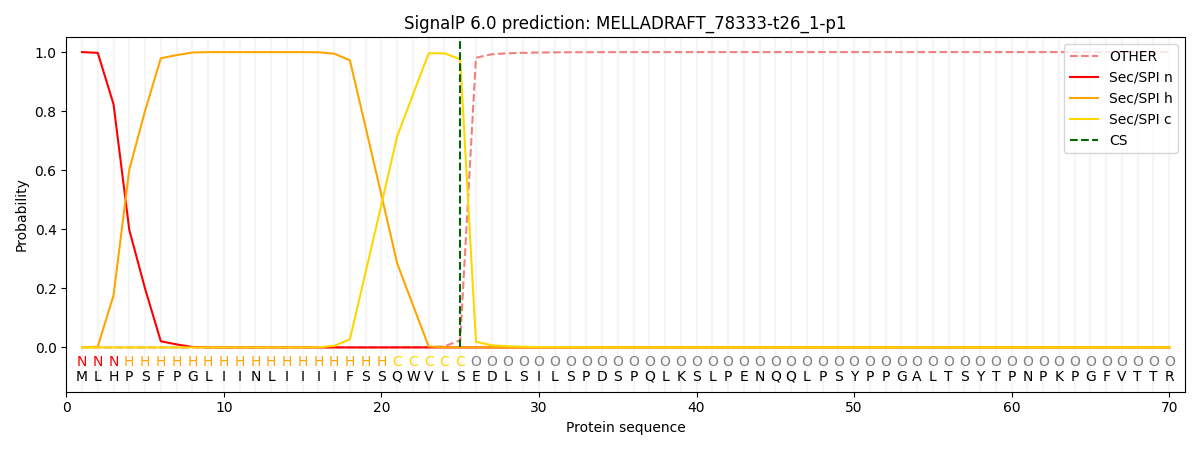
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000338 | 0.999639 | CS pos: 25-26. Pr: 0.9758 |
