You are browsing environment: FUNGIDB
CAZyme Information: MELLADRAFT_72436-t26_1-p1
You are here: Home > Sequence: MELLADRAFT_72436-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Melampsora larici-populina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Melampsoraceae; Melampsora; Melampsora larici-populina | |||||||||||
| CAZyme ID | MELLADRAFT_72436-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | family 1 polysaccharide lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 116 | 302 | 1.2e-69 | 0.983957219251337 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 3.92e-28 | 118 | 301 | 4 | 186 | Amb_all domain. |
| 226384 | PelB | 4.87e-12 | 120 | 305 | 91 | 277 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 366158 | Pec_lyase_C | 2.42e-07 | 122 | 301 | 26 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.77e-92 | 9 | 386 | 6 | 369 | |
| 1.37e-90 | 11 | 395 | 9 | 381 | |
| 7.04e-90 | 9 | 395 | 6 | 378 | |
| 3.72e-88 | 6 | 395 | 6 | 373 | |
| 4.20e-88 | 9 | 384 | 7 | 365 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.58e-78 | 24 | 369 | 5 | 335 | Pectin Lyase B [Aspergillus niger] |
|
| 3.06e-74 | 23 | 369 | 4 | 334 | Pectin Lyase A [Aspergillus niger] |
|
| 4.79e-73 | 23 | 369 | 4 | 334 | Pectin Lyase A [Aspergillus niger],1IDJ_B Pectin Lyase A [Aspergillus niger] |
|
| 4.67e-10 | 122 | 301 | 72 | 276 | Chain A, PECTATE LYASE E [Dickeya chrysanthemi] |
|
| 2.93e-06 | 126 | 278 | 128 | 301 | Structure of the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.65e-83 | 23 | 387 | 23 | 373 | Probable pectin lyase D OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=pelD PE=3 SV=1 |
|
| 6.65e-83 | 23 | 387 | 23 | 373 | Pectin lyase 2 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pel2 PE=1 SV=1 |
|
| 1.05e-81 | 12 | 387 | 12 | 373 | Probable pectin lyase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pelB PE=3 SV=2 |
|
| 4.57e-80 | 24 | 343 | 20 | 337 | Probable pectin lyase E OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pelE PE=3 SV=1 |
|
| 5.68e-80 | 24 | 395 | 25 | 378 | Probable pectin lyase F-2 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pelF-2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
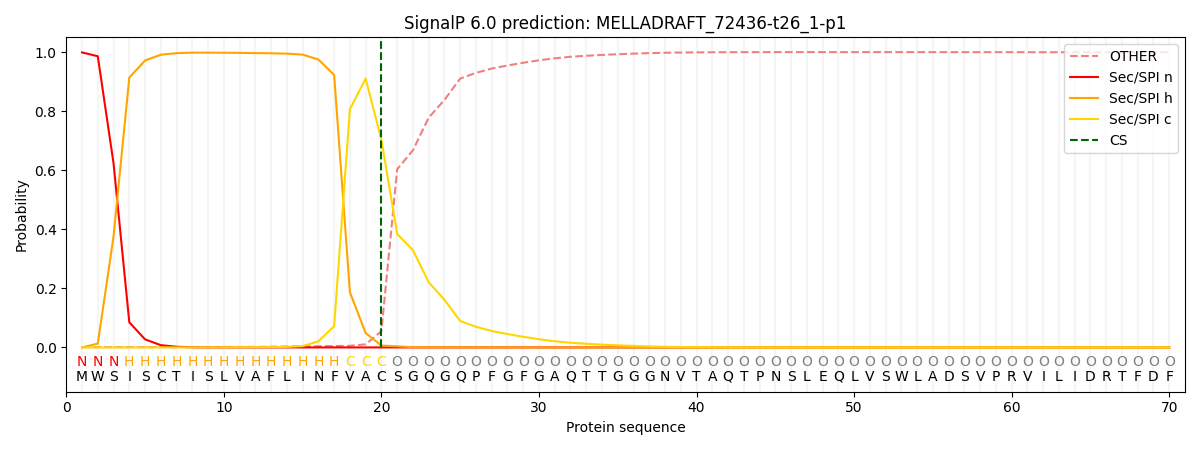
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.002521 | 0.997458 | CS pos: 20-21. Pr: 0.7032 |
