You are browsing environment: FUNGIDB
CAZyme Information: MELLADRAFT_63822-t26_1-p1
You are here: Home > Sequence: MELLADRAFT_63822-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Melampsora larici-populina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Melampsoraceae; Melampsora; Melampsora larici-populina | |||||||||||
| CAZyme ID | MELLADRAFT_63822-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | family 61 glycoside hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA9 | 100 | 273 | 5.2e-24 | 0.6454545454545455 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 410622 | LPMO_AA9 | 5.79e-26 | 70 | 289 | 38 | 216 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 9 (AA9). AA9 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs) involved in the cleavage of cellulose chains with oxidation of carbons C1 and/or C4 and C6. Activities include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54) and lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56). The family used to be called GH61 because weak endoglucanase activity had been demonstrated in some family members. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.55e-77 | 20 | 390 | 16 | 377 | |
| 7.03e-20 | 12 | 297 | 10 | 263 | |
| 1.84e-19 | 139 | 294 | 100 | 246 | |
| 3.42e-16 | 142 | 288 | 101 | 250 | |
| 3.42e-16 | 142 | 288 | 101 | 250 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.95e-10 | 147 | 290 | 84 | 226 | Extended catalytic domain of Hypocrea jecorina LPMO 9A. [Trichoderma reesei QM6a],5O2X_A Extended catalytic domain of H. jecorina LPMO9A a.k.a EG4 [Trichoderma reesei QM6a] |
|
| 5.43e-10 | 145 | 289 | 76 | 223 | Crystal structure of HiLPMO9B [Heterobasidion irregulare TC 32-1],5NNS_B Crystal structure of HiLPMO9B [Heterobasidion irregulare TC 32-1] |
|
| 5.42e-09 | 153 | 289 | 75 | 215 | Chain A, Glycoside hydrolase family 61 protein [Gelatoporia subvermispora B],7EXK_B Chain B, Glycoside hydrolase family 61 protein [Gelatoporia subvermispora B],7EXK_C Chain C, Glycoside hydrolase family 61 protein [Gelatoporia subvermispora B],7EXK_D Chain D, Glycoside hydrolase family 61 protein [Gelatoporia subvermispora B],7EXK_E Chain E, Glycoside hydrolase family 61 protein [Gelatoporia subvermispora B],7EXK_F Chain F, Glycoside hydrolase family 61 protein [Gelatoporia subvermispora B] |
|
| 7.89e-09 | 131 | 290 | 62 | 222 | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-2) [Neurospora crassa OR74A],4EIR_B Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-2) [Neurospora crassa OR74A] |
|
| 7.89e-09 | 131 | 290 | 62 | 222 | Neurospora crassa polysaccharide monooxygenase 2 high mannosylation [Neurospora crassa],5TKF_B Neurospora crassa polysaccharide monooxygenase 2 high mannosylation [Neurospora crassa],5TKF_C Neurospora crassa polysaccharide monooxygenase 2 high mannosylation [Neurospora crassa],5TKF_D Neurospora crassa polysaccharide monooxygenase 2 high mannosylation [Neurospora crassa],5TKG_A Neurospora crassa polysaccharide monooxygenase 2 resting state [Neurospora crassa],5TKG_B Neurospora crassa polysaccharide monooxygenase 2 resting state [Neurospora crassa],5TKH_A Neurospora crassa polysaccharide monooxygenase 2 ascorbate treated [Neurospora crassa],5TKH_B Neurospora crassa polysaccharide monooxygenase 2 ascorbate treated [Neurospora crassa],5TKI_B Neurospora crassa polysaccharide monooxygenase 2 resting state joint X-ray/neutron refinement [Neurospora crassa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.24e-09 | 147 | 290 | 105 | 247 | Endoglucanase-4 OS=Hypocrea jecorina OX=51453 GN=cel61a PE=1 SV=1 |
|
| 5.64e-08 | 152 | 289 | 108 | 243 | Polysaccharide monooxygenase Cel61a OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Cel61a PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
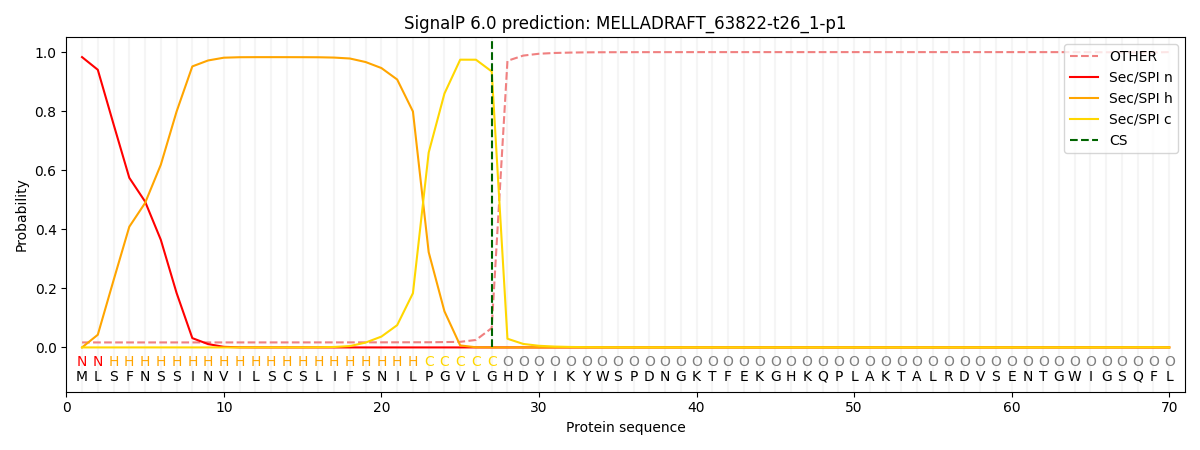
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.023184 | 0.976786 | CS pos: 27-28. Pr: 0.9345 |
