You are browsing environment: FUNGIDB
CAZyme Information: MELLADRAFT_62083-t26_1-p1
You are here: Home > Sequence: MELLADRAFT_62083-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Melampsora larici-populina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Melampsoraceae; Melampsora; Melampsora larici-populina | |||||||||||
| CAZyme ID | MELLADRAFT_62083-t26_1-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | family 5 carbohydrate esterase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 120 | 305 | 1e-29 | 0.9894179894179894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 3.77e-19 | 120 | 305 | 2 | 173 | Cutinase. |
| 223029 | PHA03264 | 0.002 | 65 | 119 | 272 | 327 | envelope glycoprotein D; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.16e-21 | 107 | 307 | 13 | 221 | |
| 1.24e-21 | 107 | 299 | 15 | 213 | |
| 1.62e-21 | 107 | 307 | 13 | 221 | |
| 5.20e-21 | 119 | 299 | 27 | 213 | |
| 5.30e-21 | 107 | 305 | 22 | 212 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.45e-18 | 119 | 288 | 2 | 178 | Chain A, Cutinase-like enzyme [Moesziomyces antarcticus],7CC4_B Chain B, Cutinase-like enzyme [Moesziomyces antarcticus],7CW1_A Chain A, Cutinase-like enzyme [Moesziomyces antarcticus],7CW1_B Chain B, Cutinase-like enzyme [Moesziomyces antarcticus] |
|
| 1.12e-06 | 120 | 305 | 17 | 193 | Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY3_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY9_A Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9],7CY9_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9] |
|
| 3.37e-06 | 120 | 310 | 31 | 212 | Chain A, CUTINASE [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.32e-17 | 120 | 309 | 35 | 217 | Carboxylesterase Culp1 OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=BQ2027_MB2006C PE=3 SV=1 |
|
| 3.32e-17 | 120 | 309 | 35 | 217 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2037 PE=3 SV=1 |
|
| 3.32e-17 | 120 | 309 | 35 | 217 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut7 PE=1 SV=1 |
|
| 2.25e-10 | 120 | 292 | 66 | 225 | Cutinase OS=Blumeria hordei OX=2867405 GN=CUT1 PE=3 SV=1 |
|
| 7.14e-10 | 120 | 288 | 25 | 213 | Cutinase 11 OS=Verticillium dahliae OX=27337 GN=VD0003_g7577 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
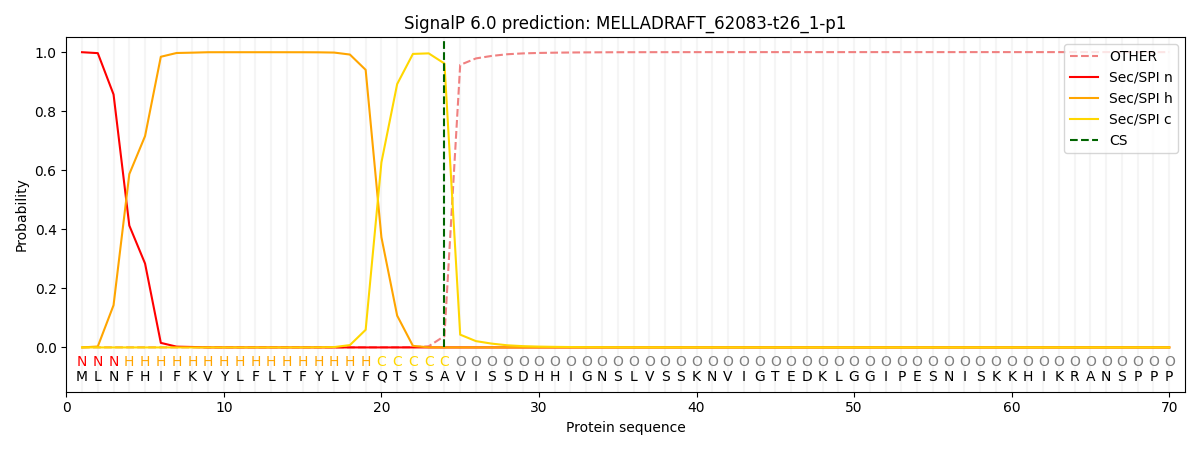
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001320 | 0.998635 | CS pos: 24-25. Pr: 0.9614 |
