You are browsing environment: FUNGIDB
CAZyme Information: MELLADRAFT_47596-t26_1-p1
You are here: Home > Sequence: MELLADRAFT_47596-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
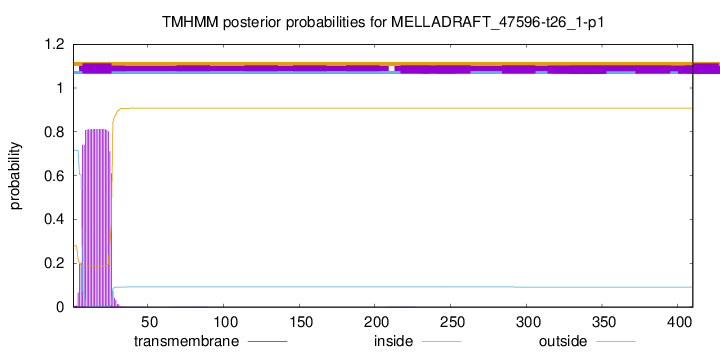
TMHMM annotations
Basic Information help
| Species | Melampsora larici-populina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Melampsoraceae; Melampsora; Melampsora larici-populina | |||||||||||
| CAZyme ID | MELLADRAFT_47596-t26_1-p1 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | family 4 carbohydrate esterase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.5.1.41:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE4 | 181 | 290 | 4.3e-22 | 0.8384615384615385 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 200576 | CE4_MrCDA_like | 3.01e-63 | 181 | 385 | 1 | 175 | Catalytic NodB homology domain of Mucor rouxii chitin deacetylase and similar proteins. This family is represented by the chitin deacetylase (MrCDA, EC 3.5.1.41) encoded from the fungus Mucor rouxii (also known as Amylomyces rouxii). MrCDA is an acidic glycoprotein with a very stringent specificity for beta1-4-linked N-acetylglucosamine homopolymers. It requires at least four residues (chitotetraose) for catalysis, and can achieve extensive deacetylation on chitin polymers. MrCDA shows high sequence similarity to Colletotrichum lindemuthianum chitin deacetylase (endo-chitin de-N-acetylase, ClCDA), which consists of a single catalytic domain similar to the deformed (beta/alpha)8 barrel fold adopted by the carbohydrate esterase 4 (CE4) superfamily, which encompasses a mononuclear metalloenzyme employing a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. The family also includes some uncharacterized eukaryotic and bacterial homologs of MrCDA. |
| 213022 | CE4_NodB_like_6s_7s | 7.54e-38 | 187 | 308 | 7 | 128 | Catalytic NodB homology domain of rhizobial NodB-like proteins. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes many rhizobial NodB chitooligosaccharide N-deacetylase (EC 3.5.1.-)-like proteins, mainly from bacteria and eukaryotes, such as chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan. All members of this family contain a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold with 6- or 7 strands. Their catalytic activity is dependent on the presence of a divalent cation, preferably cobalt or zinc, and they employ a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. Several family members show diversity both in metal ion specificities and in the residues that coordinate the metal. |
| 396211 | Polysacc_deac_1 | 1.37e-30 | 182 | 291 | 8 | 117 | Polysaccharide deacetylase. This domain is found in polysaccharide deacetylase. This family of polysaccharide deacetylases includes NodB (nodulation protein B from Rhizobium) which is a chitooligosaccharide deacetylase. It also includes chitin deacetylase from yeast, and endoxylanases which hydrolyzes glucosidic bonds in xylan. |
| 200571 | CE4_SpPgdA_BsYjeA_like | 3.43e-29 | 185 | 397 | 5 | 177 | Catalytic NodB homology domain of Streptococcus pneumoniae peptidoglycan deacetylase PgdA, Bacillus subtilis BsYjeA protein, and their bacterial homologs. This family is represented by Streptococcus pneumoniae peptidoglycan GlcNAc deacetylase (SpPgdA), a member of the carbohydrate esterase 4 (CE4) superfamily. SpPgdA protects gram-positive bacterial cell wall from host lysozymes by deacetylating peptidoglycan N-acetylglucosamine (GlcNAc) residues. It consists of three separate domains: N-terminal, middle and C-terminal (catalytic) domains. The catalytic NodB homology domain is similar to the deformed (beta/alpha)8 barrel fold adopted by other CE4 esterases, which harbors a mononuclear metalloenzyme employing a conserved His-His-Asp zinc-binding triad closely associated with conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. The enzyme is able to accept GlcNAc3 as a substrate, with the N-acetyl of the middle sugar being removed by the enzyme. This family also includes Bacillus subtilis BsYjeA protein encoded by the yjeA gene, which is one of the six polysaccharide deacetylase gene homologs (pdaA, pdaB/ybaN, yheN, yjeA, yxkH and ylxY) in the Bacillus subtilis genome. Although homology comparison shows that the BsYjeA protein contains a polysaccharide deacetylase domain, and was predicted to be a membrane-bound xylanase or a membrane-bound chitooligosaccharide deacetylase, more recent research indicates BsYjeA might be a novel non-specific secretory endonuclease which creates random nicks progressively on the two strands of dsDNA, resulting in highly distinguishable intermediates/products very different in chemical and physical compositions over time. In addition, BsYjeA shares several enzymatic properties with the well-understood DNase I endonuclease. Both enzymes are active on ssDNA and dsDNA, both generate random nicks, and both require Mg2+ or Mn2+ for hydrolytic activity. |
| 200582 | CE4_NodB_like_3 | 1.15e-26 | 187 | 313 | 7 | 133 | Catalytic NodB homology domain of uncharacterized bacterial polysaccharide deacetylases. This family includes many uncharacterized bacterial polysaccharide deacetylases. Although their biological function still remains unknown, members in this family show high sequence homology to the catalytic NodB homology domain of Streptococcus pneumoniae polysaccharide deacetylase PgdA (SpPgdA), which is an extracellular metal-dependent polysaccharide deacetylase with de-N-acetylase activity toward a hexamer of chitooligosaccharide N-acetylglucosamine, but not shorter chitooligosaccharides or a synthetic peptidoglycan tetrasaccharide. Like SpPgdA, this family is a member of the carbohydrate esterase 4 (CE4) superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.91e-61 | 91 | 410 | 33 | 313 | |
| 9.71e-57 | 91 | 410 | 27 | 306 | |
| 2.95e-51 | 80 | 409 | 54 | 335 | |
| 2.95e-51 | 80 | 409 | 54 | 335 | |
| 1.04e-50 | 75 | 409 | 53 | 344 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.64e-19 | 178 | 315 | 2 | 138 | Family 4 carbohydrate esterase from Streptomyces lividans in complex with acetate [Streptomyces lividans],2CC0_B Family 4 carbohydrate esterase from Streptomyces lividans in complex with acetate [Streptomyces lividans] |
|
| 1.80e-18 | 184 | 401 | 120 | 300 | Chain A, Predicted xylanase/chitin deacetylase [Caldanaerobacter subterraneus subsp. tengcongensis MB4] |
|
| 3.38e-17 | 169 | 401 | 226 | 416 | Structure of Streptococcus pneumoniae peptidoglycan deacetylase (SpPgdA) [Streptococcus pneumoniae R6] |
|
| 1.47e-16 | 169 | 401 | 226 | 416 | Structure of Streptococcus pneumoniae peptidoglycan deacetylase (SpPgdA) D 275 N Mutant. [Streptococcus pneumoniae R6] |
|
| 2.74e-16 | 184 | 327 | 24 | 168 | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.41e-50 | 80 | 409 | 42 | 325 | Chitin deacetylase 3 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=CDA3 PE=1 SV=3 |
|
| 1.97e-49 | 87 | 401 | 49 | 320 | Chitin deacetylase 3 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CDA3 PE=2 SV=1 |
|
| 2.75e-49 | 87 | 401 | 49 | 320 | Chitin deacetylase 3 OS=Cryptococcus neoformans var. neoformans serotype D (strain B-3501A) OX=283643 GN=CDA3 PE=3 SV=1 |
|
| 2.36e-34 | 116 | 397 | 72 | 348 | Chitin deacetylase 2 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=CDA2 PE=1 SV=1 |
|
| 4.69e-34 | 148 | 397 | 125 | 349 | Chitin deacetylase 2 OS=Cryptococcus neoformans var. neoformans serotype D (strain B-3501A) OX=283643 GN=CDA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000037 | 0.000002 |