You are browsing environment: FUNGIDB
CAZyme Information: MELLADRAFT_37509-t26_1-p1
You are here: Home > Sequence: MELLADRAFT_37509-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Melampsora larici-populina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Melampsoraceae; Melampsora; Melampsora larici-populina | |||||||||||
| CAZyme ID | MELLADRAFT_37509-t26_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | family 2 glycoside hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.146:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 52 | 511 | 2.3e-100 | 0.5159574468085106 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225789 | LacZ | 4.53e-28 | 61 | 470 | 15 | 440 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| 236657 | PRK10150 | 1.34e-22 | 50 | 322 | 3 | 279 | beta-D-glucuronidase; Provisional |
| 236673 | ebgA | 3.93e-15 | 56 | 401 | 39 | 394 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| 397120 | Glyco_hydro_2_N | 2.60e-11 | 111 | 218 | 68 | 169 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| 236548 | lacZ | 1.35e-08 | 112 | 401 | 124 | 410 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.88e-251 | 18 | 624 | 21 | 613 | |
| 1.32e-241 | 21 | 624 | 39 | 634 | |
| 8.68e-239 | 9 | 627 | 11 | 619 | |
| 3.38e-238 | 25 | 624 | 19 | 619 | |
| 3.05e-235 | 23 | 623 | 33 | 634 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.89e-117 | 32 | 621 | 12 | 587 | Chain A, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
|
| 1.98e-22 | 89 | 473 | 75 | 436 | Chain A, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_B Chain B, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_C Chain C, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_D Chain D, GH2 beta-galacturonate AqGalA [Aquimarina sp.] |
|
| 4.00e-21 | 111 | 449 | 80 | 423 | Chain A, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei] |
|
| 4.95e-20 | 111 | 447 | 87 | 426 | D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii],6D8G_B D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii] |
|
| 1.13e-19 | 111 | 447 | 79 | 418 | Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.95e-20 | 111 | 527 | 106 | 522 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
|
| 4.93e-15 | 111 | 447 | 59 | 388 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
|
| 6.03e-15 | 111 | 447 | 109 | 432 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
|
| 1.62e-12 | 112 | 447 | 58 | 389 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
|
| 7.52e-12 | 52 | 404 | 4 | 410 | Mannosylglycoprotein endo-beta-mannosidase OS=Lilium longiflorum OX=4690 GN=EBM PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
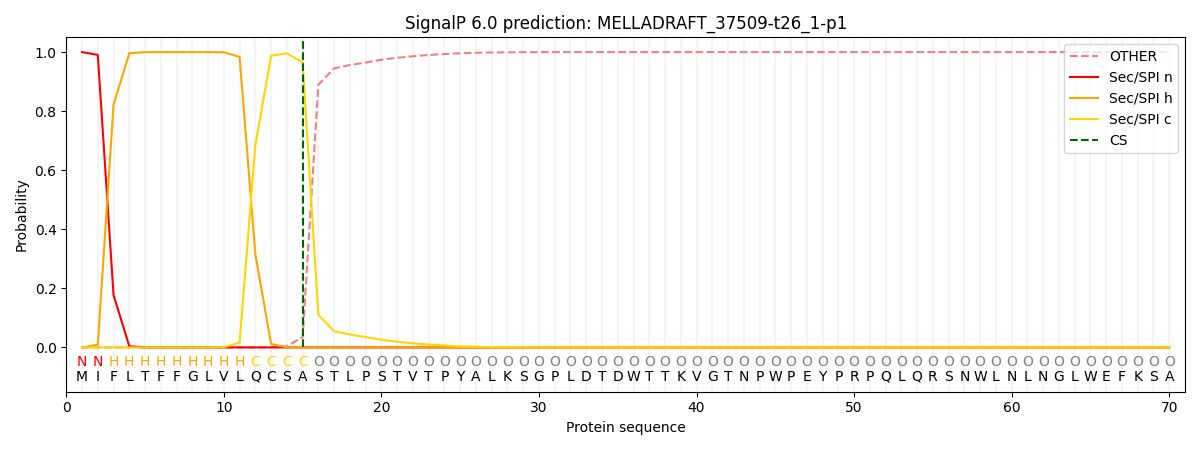
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000246 | 0.999735 | CS pos: 15-16. Pr: 0.9649 |
