You are browsing environment: FUNGIDB
CAZyme Information: MELLADRAFT_111031-t26_1-p1
You are here: Home > Sequence: MELLADRAFT_111031-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Melampsora larici-populina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Melampsoraceae; Melampsora; Melampsora larici-populina | |||||||||||
| CAZyme ID | MELLADRAFT_111031-t26_1-p1 | |||||||||||
| CAZy Family | AA5 | |||||||||||
| CAZyme Description | family 10 glycoside hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 48 | 341 | 2e-93 | 0.9471947194719472 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395262 | Glyco_hydro_10 | 1.21e-125 | 46 | 340 | 10 | 308 | Glycosyl hydrolase family 10. |
| 214750 | Glyco_10 | 2.50e-109 | 80 | 340 | 1 | 263 | Glycosyl hydrolase family 10. |
| 226217 | XynA | 2.25e-91 | 60 | 339 | 47 | 336 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.88e-101 | 37 | 333 | 27 | 328 | |
| 2.40e-94 | 60 | 344 | 53 | 338 | |
| 2.78e-93 | 60 | 344 | 57 | 342 | |
| 2.03e-92 | 60 | 344 | 63 | 350 | |
| 8.07e-92 | 49 | 343 | 48 | 343 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.45e-91 | 47 | 343 | 15 | 311 | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
|
| 6.36e-89 | 47 | 343 | 15 | 311 | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
|
| 4.24e-81 | 49 | 344 | 16 | 312 | Beta-1,4-Glycanase Cex-Cd [Cellulomonas fimi],1FH7_A Crystal Structure Of The Xylanase Cex With Xylobiose- Derived Inhibitor Deoxynojirimycin [Cellulomonas fimi],1FH8_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Isofagomine Inhibitor [Cellulomonas fimi],1FH9_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Lactam Oxime Inhibitor [Cellulomonas fimi],1FHD_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Imidazole Inhibitor [Cellulomonas fimi],1J01_A Crystal Structure Of The Xylanase Cex With Xylobiose-Derived Inhibitor Isofagomine lactam [Cellulomonas fimi],2EXO_A Crystal Structure Of The Catalytic Domain Of The Beta-1,4- Glycanase Cex From Cellulomonas Fimi [Cellulomonas fimi],2XYL_A Cellulomonas Fimi XylanaseCELLULASE COMPLEXED WITH 2-Deoxy- 2-Fluoro-Xylobiose [Cellulomonas fimi] |
|
| 4.66e-81 | 49 | 344 | 16 | 312 | Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with cellobiose-like isofagomine [Cellulomonas fimi],3CUG_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with cellotetraose-like isofagomine [Cellulomonas fimi],3CUH_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with cellotriose-like isofagomine [Cellulomonas fimi],3CUI_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with sulfur substituted beta-1,4 xylotetraose [Cellulomonas fimi],3CUJ_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with sulfur substituted beta-1,4 xylopentaose. [Cellulomonas fimi] |
|
| 3.84e-79 | 49 | 344 | 16 | 312 | Chain A, CELLULOMONAS FIMI FAMILY 10 BETA-1,4-GLYCANASE [Cellulomonas fimi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.00e-86 | 47 | 343 | 53 | 349 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
|
| 7.39e-81 | 71 | 338 | 55 | 325 | Endo-1,4-beta-xylanase OS=Agaricus bisporus OX=5341 GN=xlnA PE=2 SV=1 |
|
| 1.71e-80 | 21 | 344 | 87 | 406 | Endo-1,4-beta-xylanase A OS=Phanerodontia chrysosporium OX=2822231 GN=xynA PE=1 SV=1 |
|
| 7.65e-79 | 49 | 344 | 57 | 353 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
|
| 1.14e-72 | 47 | 344 | 95 | 397 | Endo-1,4-beta-xylanase C OS=Phanerodontia chrysosporium OX=2822231 GN=xynC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
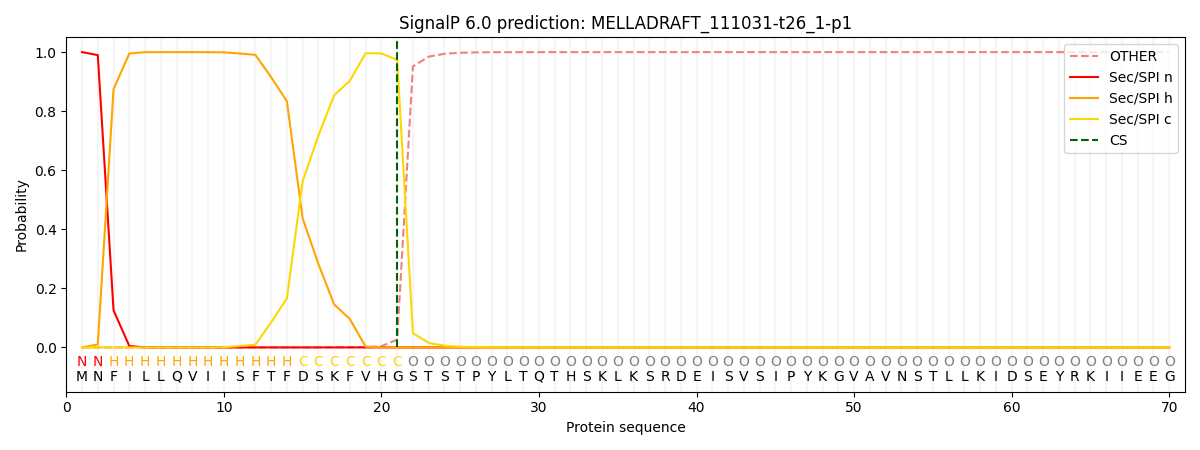
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000233 | 0.999705 | CS pos: 21-22. Pr: 0.9737 |
