You are browsing environment: FUNGIDB
CAZyme Information: M747DRAFT_297723-t43_1-p1
You are here: Home > Sequence: M747DRAFT_297723-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus niger | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus niger | |||||||||||
| CAZyme ID | M747DRAFT_297723-t43_1-p1 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | alpha-1,6-mannosyltransferase subunit | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 10 | 411 | 6.9e-87 | 0.9408740359897172 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 7.93e-37 | 60 | 379 | 44 | 365 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 215437 | PLN02816 | 4.83e-16 | 77 | 336 | 106 | 348 | mannosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 4 | 562 | 19 | 577 | |
| 0.0 | 1 | 562 | 1 | 559 | |
| 0.0 | 1 | 562 | 1 | 559 | |
| 1.31e-289 | 1 | 558 | 1 | 558 | |
| 1.31e-289 | 1 | 558 | 1 | 558 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.35e-66 | 21 | 497 | 17 | 490 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ALG12 PE=1 SV=1 |
|
| 6.01e-64 | 26 | 500 | 28 | 453 | Probable Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg12 PE=3 SV=2 |
|
| 7.62e-64 | 21 | 500 | 12 | 456 | Probable Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Drosophila melanogaster OX=7227 GN=Alg12 PE=2 SV=1 |
|
| 2.29e-62 | 10 | 500 | 14 | 460 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Mus musculus OX=10090 GN=Alg12 PE=2 SV=2 |
|
| 2.53e-60 | 9 | 485 | 12 | 453 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Homo sapiens OX=9606 GN=ALG12 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
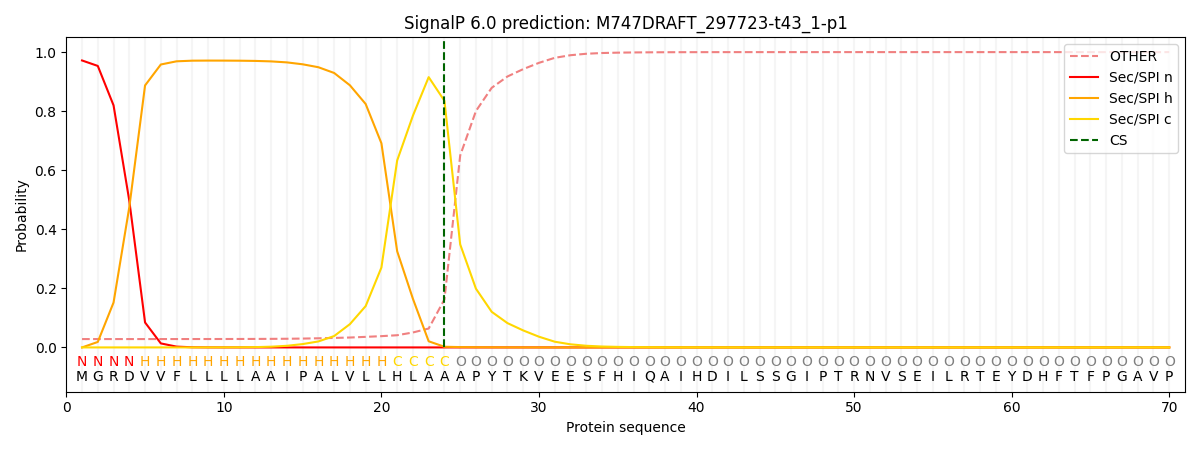
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.030553 | 0.969427 | CS pos: 24-25. Pr: 0.8351 |
TMHMM Annotations download full data without filtering help
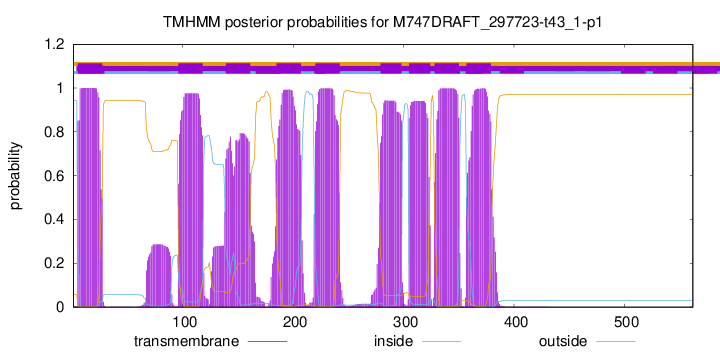
| Start | End |
|---|---|
| 5 | 27 |
| 96 | 118 |
| 139 | 161 |
| 185 | 207 |
| 220 | 242 |
| 279 | 298 |
| 305 | 324 |
| 328 | 350 |
| 357 | 379 |
