You are browsing environment: FUNGIDB
CAZyme Information: M747DRAFT_264260-t43_1-p1
You are here: Home > Sequence: M747DRAFT_264260-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
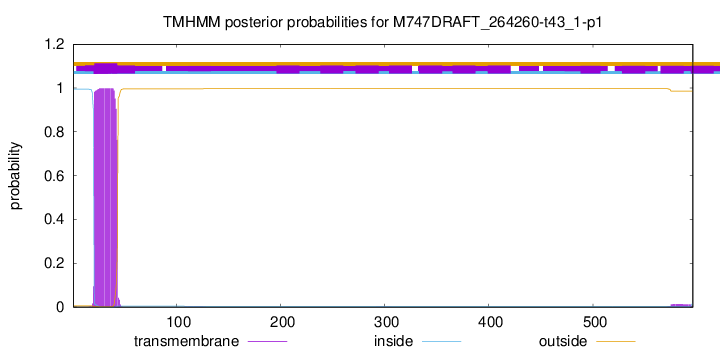
TMHMM annotations
Basic Information help
| Species | Aspergillus niger | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus niger | |||||||||||
| CAZyme ID | M747DRAFT_264260-t43_1-p1 | |||||||||||
| CAZy Family | AA9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 109 | 591 | 7.8e-93 | 0.9804469273743017 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274555 | ascorbase | 3.95e-64 | 92 | 590 | 1 | 517 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 225043 | SufI | 4.43e-64 | 97 | 591 | 38 | 443 | Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) [Cell cycle control, cell division, chromosome partitioning, Inorganic ion transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| 177843 | PLN02191 | 3.46e-59 | 91 | 594 | 22 | 544 | L-ascorbate oxidase |
| 259977 | CuRO_3_MCO_like_4 | 1.06e-58 | 417 | 595 | 2 | 165 | The third cupredoxin domain of uncharacterized multicopper oxidase. Multicopper Oxidases (MCOs) are multi-domain enzymes that are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs oxidize their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre which binds a dioxygen. The dioxygen, following the transfer of four electrons, is reduced to two molecules of water. These MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. This subfamily of MCOs is composed of three cupredoxin domains. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 215324 | PLN02604 | 2.27e-56 | 92 | 595 | 24 | 545 | oxidoreductase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 11 | 596 | 1 | 586 | |
| 2.91e-286 | 159 | 595 | 1 | 458 | |
| 2.91e-286 | 159 | 595 | 1 | 458 | |
| 5.02e-244 | 12 | 595 | 9 | 597 | |
| 1.84e-243 | 12 | 595 | 26 | 614 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.72e-58 | 92 | 594 | 2 | 477 | Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_B Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_C Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_D Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_E Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_F Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae] |
|
| 1.07e-52 | 97 | 592 | 71 | 538 | Structure of the L499M mutant of the laccase from B.aclada [Botrytis aclada] |
|
| 2.81e-52 | 97 | 592 | 71 | 538 | Crystal structure of laccase from Botrytis aclada at 1.67 A resolution [Botrytis aclada],4X4K_A Structure of laccase from Botrytis aclada with full copper content [Botrytis aclada] |
|
| 1.96e-49 | 72 | 591 | 8 | 499 | Crystal structure of a laccase-like multicopper oxidase McoG from from Aspergillus niger [Aspergillus niger] |
|
| 1.99e-49 | 72 | 591 | 9 | 500 | Crystal structure of a laccase-like multicopper oxidase McoG from Aspergillus niger bound to zinc [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.85e-62 | 91 | 590 | 23 | 490 | Iron transport multicopper oxidase FET3 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=FET3 PE=2 SV=1 |
|
| 9.56e-62 | 91 | 594 | 26 | 503 | Iron transport multicopper oxidase FET3 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=FET3 PE=3 SV=1 |
|
| 4.16e-61 | 91 | 591 | 21 | 490 | Iron transport multicopper oxidase fetC OS=Epichloe festucae (strain E2368) OX=696363 GN=fetC PE=2 SV=1 |
|
| 7.99e-61 | 99 | 591 | 30 | 508 | Iron transport multicopper oxidase FET5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FET5 PE=1 SV=1 |
|
| 1.13e-59 | 90 | 589 | 59 | 553 | Laccase-1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=LAC1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000008 | 0.000005 |