You are browsing environment: FUNGIDB
CAZyme Information: KZZ95136.1
You are here: Home > Sequence: KZZ95136.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ascosphaera apis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ascosphaeraceae; Ascosphaera; Ascosphaera apis | |||||||||||
| CAZyme ID | KZZ95136.1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | Beta-galactosidase [Source:UniProtKB/TrEMBL;Acc:A0A168BCT3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.23:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 38 | 359 | 7.2e-98 | 0.990228013029316 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396048 | Glyco_hydro_35 | 9.15e-114 | 37 | 359 | 1 | 315 | Glycosyl hydrolases family 35. |
| 166698 | PLN03059 | 3.67e-45 | 25 | 586 | 24 | 659 | beta-galactosidase; Provisional |
| 224786 | GanA | 3.89e-34 | 31 | 603 | 1 | 580 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| 396834 | Glyco_hydro_42 | 7.38e-10 | 54 | 187 | 4 | 139 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.77e-249 | 18 | 636 | 14 | 634 | |
| 2.08e-246 | 30 | 636 | 34 | 648 | |
| 3.27e-246 | 18 | 637 | 18 | 642 | |
| 8.25e-221 | 8 | 594 | 4 | 588 | |
| 1.06e-197 | 24 | 636 | 30 | 633 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.00e-117 | 35 | 612 | 21 | 578 | Chain A, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
|
| 9.77e-115 | 24 | 612 | 21 | 593 | Galactanase BT0290 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 3.10e-109 | 25 | 611 | 2 | 572 | Crystal structure of a beta-galactosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
|
| 8.88e-101 | 22 | 613 | 2 | 599 | Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_B Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_C Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_D Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THD_A Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_B Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_C Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_D Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens] |
|
| 1.62e-100 | 22 | 613 | 26 | 623 | Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_B Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_C Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_D Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WF0_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF1_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF2_A Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_B Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_C Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_D Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.29e-112 | 18 | 596 | 13 | 573 | Beta-galactosidase OS=Xanthomonas manihotis OX=43353 GN=bga PE=1 SV=1 |
|
| 1.30e-106 | 38 | 619 | 54 | 620 | Beta-galactosidase-1-like protein 2 OS=Mus musculus OX=10090 GN=Glb1l2 PE=1 SV=1 |
|
| 4.80e-105 | 19 | 611 | 30 | 613 | Beta-galactosidase-1-like protein 3 OS=Rattus norvegicus OX=10116 GN=Glb1l3 PE=2 SV=1 |
|
| 8.57e-105 | 38 | 611 | 80 | 637 | Beta-galactosidase-1-like protein 3 OS=Homo sapiens OX=9606 GN=GLB1L3 PE=2 SV=3 |
|
| 8.32e-104 | 38 | 611 | 54 | 613 | Beta-galactosidase-1-like protein 2 OS=Homo sapiens OX=9606 GN=GLB1L2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
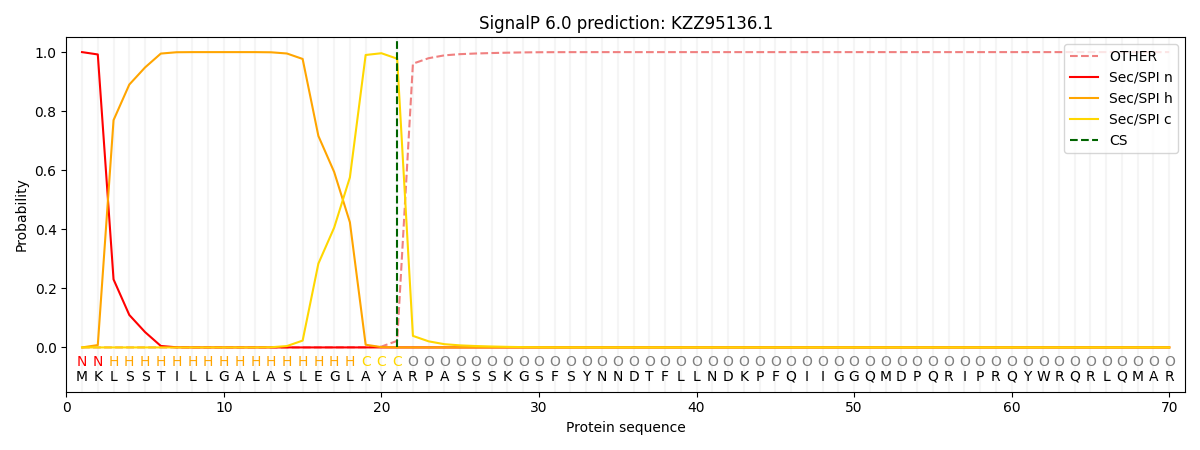
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000263 | 0.999696 | CS pos: 21-22. Pr: 0.9777 |
