You are browsing environment: FUNGIDB
CAZyme Information: KSA03536.1
You are here: Home > Sequence: KSA03536.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Debaryomyces fabryi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Debaryomyces; Debaryomyces fabryi | |||||||||||
| CAZyme ID | KSA03536.1 | |||||||||||
| CAZy Family | GT58 | |||||||||||
| CAZyme Description | GPI mannosyltransferase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT50 | 135 | 407 | 1.9e-84 | 0.9961832061068703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178434 | PLN02841 | 4.55e-73 | 9 | 407 | 11 | 415 | GPI mannosyltransferase |
| 252941 | Mannosyl_trans | 7.15e-61 | 135 | 407 | 1 | 259 | Mannosyltransferase (PIG-M). PIG-M has a DXD motif. The DXD motif is found in many glycosyltransferases that utilize nucleotide sugars. It is thought that the motif is involved in the binding of a manganese ion that is required for association of the enzymes with nucleotide sugar substrates. |
| 399599 | PIG-U | 1.44e-05 | 53 | 245 | 47 | 242 | GPI transamidase subunit PIG-U. Many eukaryotic proteins are anchored to the cell surface via glycosylphosphatidylinositol (GPI), which is posttranslationally attached to the carboxyl-terminus by GPI transamidase. The mammalian GPI transamidase is a complex of at least four subunits, GPI8, GAA1, PIG-S, and PIG-T. PIG-U is thought to represent a fifth subunit in this complex and may be involved in the recognition of either the GPI attachment signal or the lipid portion of GPI. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.01e-265 | 1 | 412 | 1 | 412 | |
| 3.54e-170 | 3 | 408 | 1 | 413 | |
| 3.54e-170 | 3 | 408 | 1 | 413 | |
| 3.54e-170 | 3 | 408 | 1 | 413 | |
| 3.54e-170 | 3 | 408 | 1 | 413 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.13e-266 | 1 | 412 | 1 | 412 | GPI mannosyltransferase 1 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=GPI14 PE=3 SV=2 |
|
| 1.24e-166 | 5 | 410 | 3 | 398 | GPI mannosyltransferase 1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=GPI14 PE=3 SV=1 |
|
| 1.02e-105 | 11 | 410 | 10 | 389 | GPI mannosyltransferase 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GPI14 PE=1 SV=2 |
|
| 9.41e-103 | 12 | 409 | 16 | 407 | GPI mannosyltransferase 1 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GPI14 PE=3 SV=1 |
|
| 5.66e-98 | 1 | 409 | 1 | 395 | GPI mannosyltransferase 1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=GPI14 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999888 | 0.000111 |
TMHMM Annotations download full data without filtering help
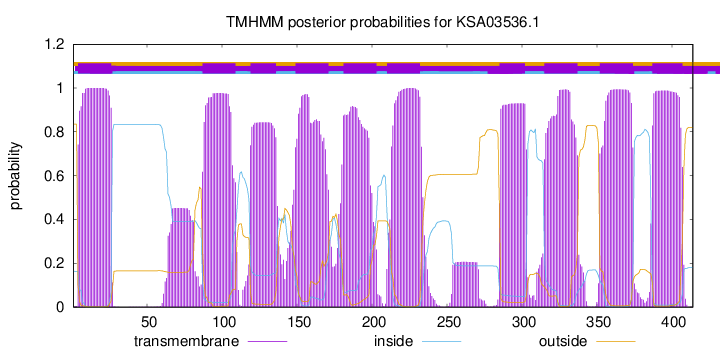
| Start | End |
|---|---|
| 4 | 26 |
| 87 | 109 |
| 119 | 136 |
| 149 | 171 |
| 181 | 203 |
| 210 | 232 |
| 285 | 302 |
| 315 | 337 |
| 352 | 374 |
| 387 | 407 |
