You are browsing environment: FUNGIDB
CAZyme Information: KNZ61589.1
You are here: Home > Sequence: KNZ61589.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia sorghi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia sorghi | |||||||||||
| CAZyme ID | KNZ61589.1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | alpha-1,2-Mannosidase [Source:UniProtKB/TrEMBL;Acc:A0A0L6VLK6] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | - | 3.2.1.113:6 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 253 | 477 | 1.4e-60 | 0.547085201793722 |
| GH47 | 64 | 253 | 3.2e-58 | 0.3789237668161435 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396217 | Glyco_hydro_47 | 4.43e-133 | 64 | 476 | 3 | 451 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| 240427 | PTZ00470 | 2.03e-70 | 50 | 480 | 71 | 520 | glycoside hydrolase family 47 protein; Provisional |
| 238300 | PA | 2.44e-12 | 779 | 869 | 41 | 117 | PA: Protease-associated (PA) domain. The PA domain is an insert domain in a diverse fraction of proteases. The significance of the PA domain to many of the proteins in which it is inserted is undetermined. It may be a protein-protein interaction domain. At peptidase active sites, the PA domain may participate in substrate binding and/or promoting conformational changes, which influence the stability and accessibility of the site to substrate. Proteins into which the PA domain is inserted include the following: i) various signal peptide peptidases including, hSPPL2a and 2b which catalyze the intramembrane proteolysis of tumor necrosis factor alpha, ii) various proteins containing a C3H2C3 RING finger including, Arabidopsis ReMembR-H2 protein and various E3 ubiquitin ligases such as human GRAIL (gene related to anergy in lymphocytes), iii) EDEM3 (ER-degradation-enhancing mannosidase-like 3 protein), iv) various plant vacuolar sorting receptors such as Pisum sativum BP-80, v) glutamate carboxypeptidase II (GCPII), vi) yeast aminopeptidase Y, vii) Vibrio metschnikovii VapT, a sodium dodecyl sulfate (SDS) resistant extracellular alkaline serine protease, viii) lactocepin (a cell envelope-associated protease from Lactobacillus paracasei subsp. paracasei NCDO 151), ix) various subtilisin-like proteases such as melon Cucumisin, and x) human TfR (transferrin receptor) 1 and 2. |
| 239048 | PA_C5a_like | 1.71e-09 | 781 | 827 | 44 | 90 | PA_C5a_like: Protease-associated domain containing proteins like Streptococcus pyogenes C5a peptidase. This group contains various PA domain-containing proteins similar to S. pyogenes C5a, including, i) Vpr, a minor extracellular serine protease from Bacillus subtilis, ii) a large molecular mass collagenolytic protease from Geobacillus collagenovorans MO-1, and iii) PrtS, a cell envelope protease from Streptococcus thermophilus CNRZ 385. Proteins in this group belong to the peptidase S8 family. C5a peptidase is a cell surface serine protease which specifically inactivates C5a [a chemotactic peptide, which attracts polymorphonuclear leukocytes (PMNs)], by cleaving it to release a 7-residue carboxy-terminal fragment which contains the PMN binding site. The significance of the PA domain to these proteins has not been ascertained. It may be a protein-protein interaction domain. At peptidase active sites, the PA domain may participate in substrate binding and/or promoting conformational changes, which influence the stability and accessibility of the site to substrate. |
| 239041 | PA_EDEM3_like | 1.08e-08 | 781 | 820 | 37 | 76 | PA_EDEM3_like: protease associated domain (PA) domain-containing EDEM3-like proteins. This group contains various PA domain-containing proteins similar to mouse EDEM3 (ER-degradation-enhancing mannosidase-like 3 protein). EDEM3 contains a region, similar to Class I alpha-mannosidases (gylcosyl hydrolase family 47), N-terminal to the PA domain. EDEM3 accelerates glycoprotein ERAD (ER-associated degradation). In transfected mammalian cells, overexpression of EDEM3 enhances the mannose trimming from the N-glycans, of a model misfolded protein [alpha1-antitrypsin null (Hong Kong)] as well as, from total glycoproteins. Mannose trimming appears to be involved in the selection of ERAD substrates. EDEM3 has a different specificity of trimming than ER alpha-mannosidase 1. The significance of the PA domain to EDEM3 has not been ascertained. It may be a protein-protein interaction domain. At peptidase active sites, the PA domain may participate in substrate binding and/or promoting conformational changes, which influence the stability and accessibility of the site to substrate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.58e-179 | 48 | 827 | 45 | 787 | |
| 1.41e-175 | 48 | 827 | 45 | 787 | |
| 5.09e-162 | 43 | 828 | 39 | 831 | |
| 1.22e-160 | 46 | 872 | 26 | 830 | |
| 3.92e-160 | 50 | 921 | 37 | 958 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.32e-36 | 59 | 478 | 9 | 451 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
|
| 4.78e-36 | 59 | 478 | 14 | 456 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase [Homo sapiens] |
|
| 4.94e-36 | 59 | 478 | 14 | 456 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With 1-Deoxymannojirimycin [Homo sapiens],1FO3_A Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With Kifunensine [Homo sapiens] |
|
| 1.53e-35 | 59 | 478 | 92 | 534 | Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue [Homo sapiens] |
|
| 2.02e-35 | 59 | 478 | 9 | 451 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens],5KK7_B Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.94e-129 | 27 | 496 | 4 | 506 | ER degradation-enhancing alpha-mannosidase-like protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mnl1 PE=3 SV=2 |
|
| 5.49e-123 | 49 | 478 | 119 | 581 | ER degradation-enhancing alpha-mannosidase-like protein 1 OS=Mus musculus OX=10090 GN=Edem1 PE=1 SV=1 |
|
| 6.33e-123 | 49 | 478 | 124 | 586 | ER degradation-enhancing alpha-mannosidase-like protein 1 OS=Homo sapiens OX=9606 GN=EDEM1 PE=1 SV=1 |
|
| 1.69e-105 | 27 | 481 | 9 | 476 | Alpha-mannosidase I MNS4 OS=Arabidopsis thaliana OX=3702 GN=MNS4 PE=1 SV=1 |
|
| 1.77e-104 | 30 | 485 | 8 | 509 | ER degradation-enhancing alpha-mannosidase-like protein 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNL1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
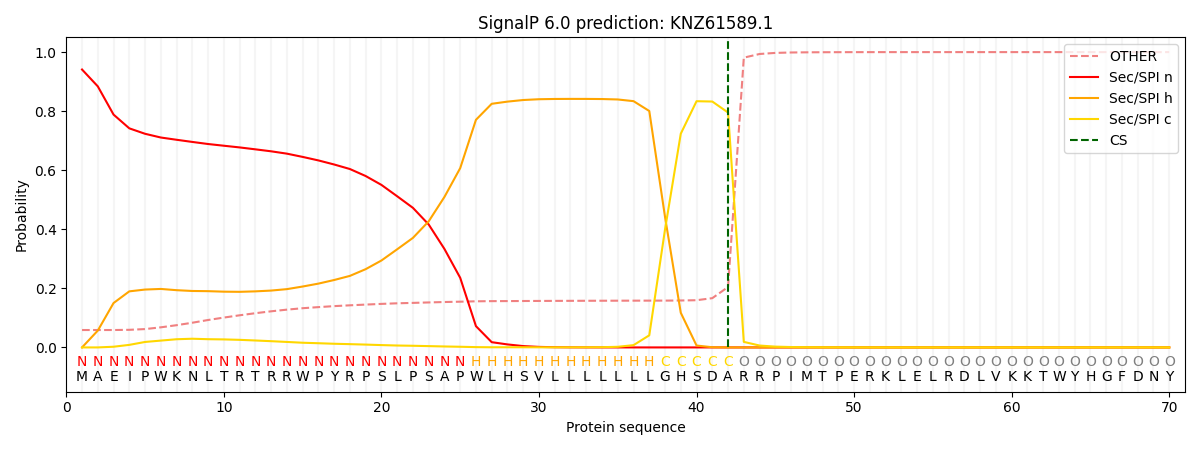
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.060106 | 0.939863 | CS pos: 42-43. Pr: 0.7954 |
