You are browsing environment: FUNGIDB
CAZyme Information: KNZ51833.1
You are here: Home > Sequence: KNZ51833.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia sorghi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia sorghi | |||||||||||
| CAZyme ID | KNZ51833.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.135:38 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT43 | 212 | 399 | 1.4e-21 | 0.9339622641509434 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397440 | Glyco_transf_43 | 8.05e-27 | 212 | 399 | 9 | 198 | Glycosyltransferase family 43. |
| 132995 | GlcAT-I | 6.25e-25 | 212 | 399 | 30 | 218 | Beta1,3-glucuronyltransferase I (GlcAT-I) is involved in the initial steps of proteoglycan synthesis. Beta1,3-glucuronyltransferase I (GlcAT-I) domain; GlcAT-I is a Key enzyme involved in the initial steps of proteoglycan synthesis. GlcAT-I catalyzes the transfer of a glucuronic acid moiety from the uridine diphosphate-glucuronic acid (UDP-GlcUA) to the common linkage region of trisaccharide Gal-beta-(1-3)-Gal-beta-(1-4)-Xyl of proteoglycans. The enzyme has two subdomains that bind the donor and acceptor substrate separately. The active site is located at the cleft between both subdomains in which the trisaccharide molecule is oriented perpendicular to the UDP. This family has been classified as Glycosyltransferase family 43 (GT-43). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.12e-98 | 128 | 408 | 16 | 288 | |
| 2.01e-14 | 214 | 400 | 21 | 217 | |
| 5.89e-13 | 210 | 400 | 110 | 304 | |
| 6.66e-13 | 210 | 400 | 62 | 259 | |
| 1.40e-12 | 210 | 400 | 107 | 301 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.94e-14 | 210 | 400 | 30 | 224 | Crystal Structure of Human GlcAT-S Apo Form [Homo sapiens],2D0J_B Crystal Structure of Human GlcAT-S Apo Form [Homo sapiens],2D0J_C Crystal Structure of Human GlcAT-S Apo Form [Homo sapiens],2D0J_D Crystal Structure of Human GlcAT-S Apo Form [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.48e-13 | 210 | 400 | 107 | 301 | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 OS=Homo sapiens OX=9606 GN=B3GAT2 PE=1 SV=2 |
|
| 2.61e-13 | 210 | 400 | 113 | 307 | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 OS=Canis lupus familiaris OX=9615 GN=B3GAT2 PE=2 SV=1 |
|
| 8.38e-09 | 214 | 412 | 128 | 335 | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase sqv-8 OS=Caenorhabditis elegans OX=6239 GN=sqv-8 PE=1 SV=1 |
|
| 2.63e-07 | 214 | 401 | 262 | 449 | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase P OS=Drosophila melanogaster OX=7227 GN=GlcAT-P PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
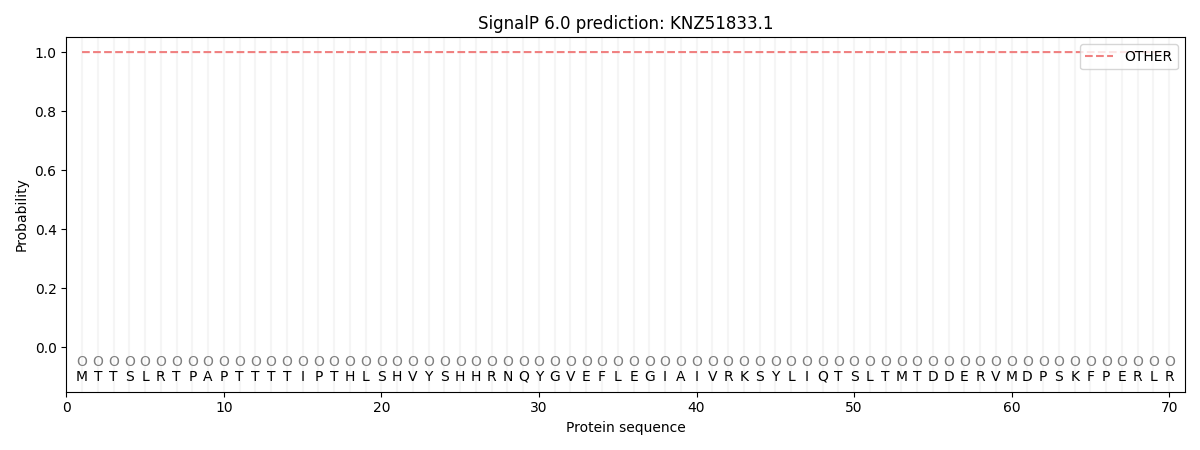
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000062 | 0.000001 |
