You are browsing environment: FUNGIDB
CAZyme Information: KNZ48999.1
You are here: Home > Sequence: KNZ48999.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia sorghi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia sorghi | |||||||||||
| CAZyme ID | KNZ48999.1 | |||||||||||
| CAZy Family | GH131 | |||||||||||
| CAZyme Description | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase [Source:UniProtKB/TrEMBL;Acc:A0A0L6UKE5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.256:6 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT59 | 33 | 460 | 5.8e-109 | 0.9925742574257426 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398537 | DIE2_ALG10 | 3.58e-137 | 33 | 460 | 1 | 383 | DIE2/ALG10 family. The ALG10 protein from Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum. This protein has been characterized in rat as potassium channel regulator 1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.66e-85 | 16 | 515 | 10 | 483 | |
| 1.24e-82 | 16 | 515 | 10 | 463 | |
| 2.91e-82 | 13 | 515 | 8 | 507 | |
| 3.78e-82 | 17 | 515 | 15 | 504 | |
| 7.88e-74 | 46 | 515 | 39 | 470 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.31e-66 | 12 | 515 | 10 | 474 | Putative Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Rattus norvegicus OX=10116 GN=Alg10b PE=1 SV=1 |
|
| 7.05e-66 | 12 | 515 | 10 | 474 | Putative Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Mus musculus OX=10090 GN=Alg10b PE=1 SV=1 |
|
| 1.26e-63 | 29 | 515 | 21 | 509 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Arabidopsis thaliana OX=3702 GN=ALG10 PE=1 SV=1 |
|
| 8.30e-62 | 12 | 515 | 10 | 473 | Putative Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Homo sapiens OX=9606 GN=ALG10B PE=1 SV=2 |
|
| 2.27e-61 | 12 | 515 | 10 | 473 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Homo sapiens OX=9606 GN=ALG10 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.993423 | 0.006593 |
TMHMM Annotations download full data without filtering help
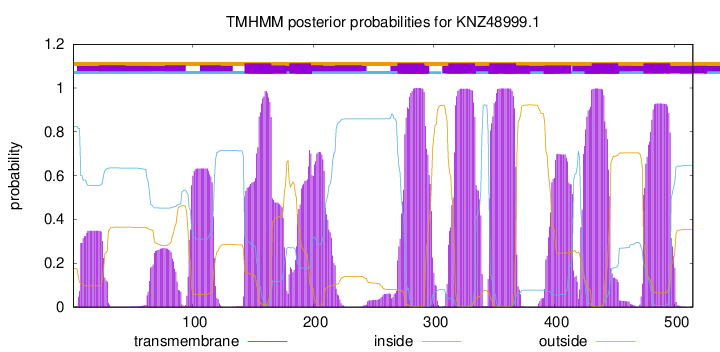
| Start | End |
|---|---|
| 143 | 165 |
| 180 | 198 |
| 270 | 292 |
| 312 | 334 |
| 346 | 368 |
| 431 | 453 |
| 474 | 496 |
